ABSTRACT
INTRODUCTION The consequences of de novo balanced structural chromosome aberrations diagnosed antenatally are unpredictable, and, as a result, they introduce uncertainty into genetic counseling decisions.
OBJECTIVE Describe de novo balanced structural aberrations present at antenatal diagnosis in samples from pregnant women in five Latin American countries and determine their effect on carrier individuals.
METHODS This was a retrospective observational study based on analysis of 109,011 antenatal tests conducted from January 1981 to December 2016 in Cuba, Uruguay, Costa Rica, Mexico, and Colombia. Thirteen cytogenetic laboratories provided information that included the cases analyzed during the study period; number of de novo balanced structural aberrations diagnosed antenatally; number of diagnoses with de novo balanced structural aberrations that resulted in termination of pregnancy; detailed descriptions of the karyotypes of de novo balanced structural aberration carriers, and descriptions of the form of diagnosis, including types of samples used (amniotic fluid, chorionic villus or fetal blood). Each laboratory also provided pathology reports and genetic counseling at time of diagnosis. Postnatal followup for pregnancies carried to term continued for at least two years.
RESULTS Of the 109,011 antenatal tests studied, 72 (0.07%) showed de novo balanced structural aberrations. These events primarily involved chromosomes 1, 2, 7, 14, 18, and 20. Of the 79 breakpoints identified, the most common were 5p15.3, 7q11.2, 7q22, and 14q24. We identified three breakpoints corresponding to 3.8% (3q13.1, 3q13.2, and 9p12) that were not reported in other studies of de novo balanced structural aberrations diagnosed antenatally in patients from other geographic regions or in studies of chromosomal fragile sites. Two of these breakpoints (3q13.1 and 3q13.2) were associated with high risk of phenotypic abnormalities. Information on antenatal or postnatal followup was available for 62 (86%) of de novo balanced structural aberration carriers; of the 44 carriers with postnatal followup, 10 had phenotypic abnormalities.
CONCLUSIONS Three new de novo breakpoints were identified, presumably related to genetic admixture characteristics in Latin America. Since some diseases associated with de novo balanced structural aberrations detected antenatally have a late onset, followup for at least two years is recommended for carriers of these aberrations. The information in this study is useful in genetic counseling for pregnant women in Latin America.
KEYWORDS Antenatal diagnosis, prenatal diagnosis, antenatal screening, chromosomal aberrations, chromosomal breakpoints, pregnancy, Colombia, Costa Rica, Cuba, Mexico, Uruguay, Latin America
INTRODUCTION
Structural chromosome aberrations usually involve changes in the linear sequence of genes due to loss, gain, or reallocation of segments, but the number of chromosomes generally remains constant. Structural changes may involve one or both chromatids. Chromatid-type aberrations differ from chromosome-type aberrations.[1] Structural chromosome rearrangements are considered balanced if disomy is maintained for all autosomes and a normal complement of sex chromatin is present, even if the positions of homologous segments on the chromosomes have been changed.
Structural chromosome rearrangements may arise at different stages of human development. When rearrangements occur in the postnatal period, they are referred to as acquired and may cause malignant diseases.[1]
IMPORTANCE These results will be useful in antenatal genetic counseling for pregnant women in Latin America and support long-term postnatal followup in patients with de novo balanced structural aberrations.
If a balanced chromosome rearrangement (BCR) is inherited, risk for physical or mental abnormalities in the newborn is low. However, if the aberration occurs de novo, i.e., if neither parent is a carrier, risk of a genetic disease or phenotypic abnormalities increases. The consequences of de novo BCRs diagnosed antenatally are especially difficult to predict. Microarray-based comparative genomic hybridization in antenatal diagnosis has shown that de novo BCRs can cause submicroscopic chromosomal imbalances—deletions, duplications and variations in the number of copies—that could alter gene dosage, inactivate genes susceptible to uniparental disomy, inactivate dominant genes (with the resulting expression of recessive deleterious genes in the homologous chromosome) and modulate gene expression around breakpoints.[2–4]
In 1991, Warburton studied 377,000 antenatal diagnoses in the USA and Canada to determine chromosomal distribution of breakpoints in de novo BCRs and to compare them with known breakpoints often associated with fragile sites in humans.[5] In this pioneer study, Warburton determined that de novo Robertsonian translocations carried a low risk for development of abnormalities (3%), while de novo inversions and translocations carried a higher risk, 9.4% and 6.1%, respectively.[5]
The demographic history of populations—with their fluctuating composition, size, and structure—may affect whole-genome patterns of variation. In addition, evolutionary processes—natural selection, mutation and genetic recombination—have affected variation in specific regions.[6]
In 2009, Giardino gathered data from 269,371 antenatal cytogenetic studies from 29 laboratories in Italy. Even though there was no pregnancy followup, she identified the chromosomes and breakpoints most involved in de novo BCR events.[7] Other studies have been conducted on prevalence of de novo BCRs in antenatal diagnosis in Asia and Australia, but the number of cases studied was smaller.[8–12]
In a study of individuals from 52 populations in Africa, Europe, the Americas, the Middle East, Asia, and Oceania, Rosenberg found that only 3%–5% of genetic variation was among populations, the majority of variation being within populations.[13]
In Latin America and the Caribbean, little has been published on de novo BCRs and their effects on neonatal phenotype by breakpoint involved.[14–17] Due to lack of information in the Region, genetic counseling for de novo BCRs in antenatal diagnosis is based on data compiled by other authors in populations that are geographically very distant and have some differences in their genome compared to our populations.[13] It is unknown what happens in relation to de novo BCRs in our Latin American populations, which are characterized primarily by a high admixture of Caucasian, African and Amerindian descent, with a lower percentage of Asian influence.[18–22] This study’s objective was to identify de novo BCRs detected antenatally in samples from five Latin American countries, and to determine their effect on individual carriers.
METHODS
Study type and population A retrospective observational study was conducted based on database information from eight cytogenetic laboratories in Cuba, one in Costa Rica, one in Colombia, two in Mexico, and one in Uruguay. Data were compiled from 109,011 antenatal tests done January 1981 through December 2016. An aberration was considered a de novo BCR if neither parent in the study had the chromosomal aberration detected in the fetus. There were 72 cases with de novo BCRs in the sample.
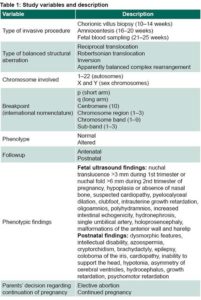
Data collection All participating laboratories were sent a questionnaire regarding the total number of cases tested, the cases in which a de novo BCR was detected, and the antenatal diagnostic method used (amniocentesis, chorionic villus biopsy or fetal blood sampling). Table 1 lists and describes the study variables. Laboratories submitted anatomic pathology reports of fetal testing in cases of pregnancy termination after detection of a de novo BCR.
Antenatal cytogenetic diagnosis Cultures and chromosome preparations were conducted according to the AGT Cytogenetics Laboratory Manual protocol[23] adapted to each laboratory’s conditions. GTG or QFQ bands were used for chromosomal antenatal diagnosis. Chorionic villus (CV) biopsy, amniocentesis and fetal blood sampling (FBS) were carried out in essentially the same way in all participating laboratories, according to procedures reported.[23]
Followup In fetuses with de novo BCRs detected antenatally whose parents decided to continue the pregnancy (49/72, 68%), a minimum two-year postnatal followup was performed, even though this was not mandatory for inclusion in the study. This two-year followup period was selected because neurodevelopmental or motor disorders are more easily detected at age 2 years, and dysmorphic features become more evident. In addition, if there is any kind of metabolic disorder, symptoms will have already appeared in most cases. Followup was performed by clinical genetics specialists. Followup was not possible in some cases because the mother did not attend the genetic followup appointment, parents emigrated or other undetermined reasons. Phenotypic abnormalities were recorded in databases (municipal, provincial, or national) and/or in medical records.
Analysis Breakpoints were not determined by advanced molecular techniques (FISH and/or microarray) due to their high cost or unavailability. The breakpoint was determined in translocations and inversions by studying the chromosome ideogram with 450 bands. Due to their low risk, Robertsonian translocations were not included in assessing risk associated with breakpoints. The International System for Human Cytogenetic Nomenclature 2005 was used to define chromosome formulae and specify sites within chromosomes: The centromere is assigned the number 10, with numbers assigned to regions on the short and long arms (p and q, respectively), getting larger in proportion to distance from the centromere. There are a limited number of bands in each region; if these bands are subdivided into sub-bands, a decimal point is added after the band designation, followed by the number of each sub-band. For example, 1p31.3 means: chromosome 1, short arm, region 3, band 1, and sub-band 3.[24] The resolution level of the analyzed metaphases is generally 450 bands.
While use of ultrasound findings can introduce bias in research on de novo BCRs, this study included cases in which some type of abnormality was detected antenatally by fetal ultrasound. Antenatal abnormalities detected by ultrasound have been shown to be important markers in detecting certain chromosome defects and, as a result, ultrasound findings are valuable in genetic counseling.[25]
The frequency distributions of detected aberrations (reciprocal translocations, Robertsonian translocations, inversions and complex rearrangements) were obtained for the different sample types.
Ethics Genetic counseling was provided to couples in all participating sites and written informed consent was obtained for specimen collection. Once antenatal diagnosis was made, all unused samples were discarded. Couples were informed of the results of cytogenetic diagnosis. In all laboratories, each patient was assigned a code in the database and anonymity was maintained during data processing. The study was approved by the ethics committees of all participating institutions.
RESULTS
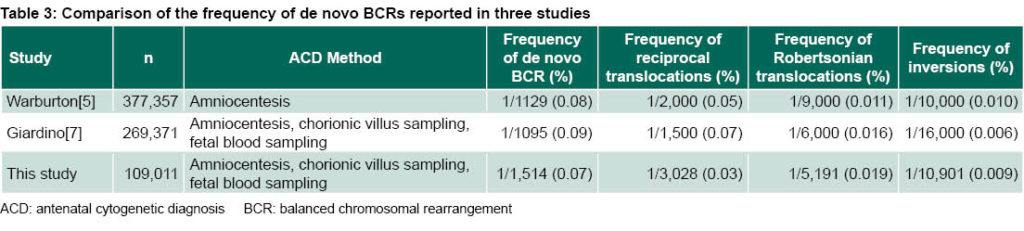
Frequency of de novo BCR Table 2 summarizes data provided by the 13 participating laboratories, from 97,790 amniotic fluid samples, 10,623 chorionic villus samples and 598 umbilical cord blood samples for a total of 109,011 antenatal diagnoses. There were 72 de novo BCRs detected, for a frequency of 1/1535 (0.07%) diagnosed cases. The laboratories detected 36 reciprocal translocations (50%), 21 Robertsonian translocations (29.1%), 10 inversions (13.9%) and five complex rearrangements (6.9%).

Amniocentesis was used for diagnosis in 89.7% (97,790/109,011) of cases (Table 1); as a result, 95.8% (69/72) of de novo BCRs were detected using this form of antenatal diagnosis.
Three mosaics were detected in reciprocal translocations, two mosaics in Robertsonian translocations, and one mosaic in cases with inversion. Of five cases with complex rearrangements, three inherited structural aberrations and one de novo aberration were found. Two cases had rearrangements involving three or more chromosomes (Data available in Appendix 1 at www.mediccreview.org/antenatal-diagnosis-of-de-novo-balanced -structural-chromosome-aberrations-in-latin-america).
As seen in Table 3, the frequency of de novo BCRs in this study is slightly different than seen in other studies with a much greater number of cases.[5,7] In general, de novo BCRs appear less frequently in our study.
Distribution of de novo BCR in chromosomes Figure 1 shows the chromosomes most commonly involved in de novo BCRs (1, 2, 7, 14, 18, and 20). Chromosomes 10, 11, 15, 17, 19, and X were very rarely involved in rearrangements. Chromosome Y did not appear in any of the reported rearrangements.
Breakpoint distribution by chromosome for reciprocal translocations and inversions in the long and short arms of the chromosomes is also shown. Many of these breakpoints coincide with those reported by Warburton[5] and Giardino.[7] However, we found breakpoints not reported in these previous studies.
Distribution of de novo BCR frequency None of the reciprocal translocations were found more than once in this study. The common translocations reported by Warburton and Giardino, t(11;22)(q23;qll) and t(18;21)(pll;qll), also appeared in our study.
Ten cases were found with inversions in which chromosomes 2, 3, and 19 were involved twice. In this study, the chromosome inversion 2(p11q13) commonly appears, even though it has been primarily described as familial.[26,27]
Breakpoints most commonly found in our study that coincide with other studies[5,7] were: 5p15.3, 7q11.2, 7q22, and 14q24. Other breakpoints that appeared once in our study and that were commonly reported by Warburton and Giardino were: 1p13.2, 2p15, 2p21, 2q21.3, 3q21, 6q21, 7p15.3, 7p22, 8q24.1, 9p13.3, 11p15, 11q23, 11q13, 21q22.3, and 22q11 (Figure 1).

Breakpoints 1q25 and 7q22 were found three times in our study. Other recurring breakpoints were: 2q21.3, 4q35, 5p15.3, 7q11.2, 14q24, 20p12, and 20q13.1, each occurring twice (Figure 1).
Of the 79 breakpoints we found, 8 (10%) had not been published previously in studies on de novo BCR in antenatal diagnoses. These were: 2p22, 3p24, 3q13.1, 3q13.2, 5p11, 8q21.1, 9p12, and 12q23.[5,7,9–12,28–44 ] (Figure 1).
Followup Antenatal or postnatal followup was successfully conducted in 62 (86%) of the 72 de novo BCR carriers. In 44 carriers, followup lasted at least two years. Table 4 shows the cases of de novo BCR carriers who had some phenotypic abnormality.
Neurodevelopmental disorders were the predominant abnormal phenotypic traits in cases assessed postnatally. There were two individuals who exhibited no mental disability despite having complex rearrangements in more than three chromosomes.
DISCUSSION
Reciprocal translocations constitute the biggest difference between our results and those of Warburton[5] and Giardino.[7] This kind of defect was not detected antenatally by chorionic villus sampling, and our result could be explained by low quality of GTG bands from using acetic acid in sample processing, which sometimes made bands difficult to distinguish and could have contributed to failure to detect this kind of rearrangement.

This method was applied in 9.7% of cases in our study and in 12% of cases in Giardino’s, which detected 8% of reciprocal translocations through chorionic villus sampling.[7]
Breakpoint distributions and the chromosomes involved do not occur randomly in these de novo rearrangements.[5,7] Recurring breakpoints have been found in AT-rich palindromic regions. Genome architecture is also known to predispose certain chromosomes to structural rearrangements, due to their spatial disposition within the cell nucleus.[45,46]
We found many reported breakpoints in other antenatally diagnosed de novo BCR studies, but we also found eight breakpoints that previous studies did not identify.[5,7, 9–12,
30–44] All those studies primarily included Caucasian and Asian populations, and the majority, as in this study, used cytogenetic banding methods to analyze breakpoints.
In postnatal studies, many of these de novo BCRs were examined using more sophisticated molecular techniques, such as whole-genome high-resolution array, which includes matrix-based comparative genomic hybridization or single-nucleotide polymorphism.[3,4,47] These methods can reveal cryptic chromosome imbalances, but they are used in postnatal studies in individuals with an abnormal phenotype. This introduces a bias into breakpoint analysis because samples come from individuals with phenotypic traits (mental disability, dysmorphic features, delayed growth, etc.) that signal possible chromosome rearrangement. Therefore, such cases are not included in our comparisons.
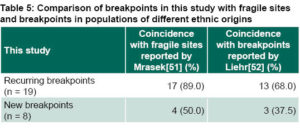
Even though the origin of a chromosome aberration is much more complex than the simple association with fragile sites inside the genome,[48–50] it is indisputable that these fragile sites are prone to formation of chromosome aberrations, primarily de novo. Fragile sites are specific loci that are prone to forming gaps and breaks on metaphase chromosomes due to partial inhibition of DNA synthesis.[51] Table 5 shows a comparison of breakpoints we found and fragile sites reported in recent international studies.[51,52] Mrasek reported 230 fragile sites induced by aphidicolin, a substance that partially inhibits DNA synthesis, in human chromosomes.[51] Liehr demonstrated that approximately 71% of breakpoints found in Caucasian individuals, studied because they had balanced aberrations, are co-located in these fragile sites, and he determined that these regions prone to chromosome breaks play an important role in formation of structural chromosome rearrangements.[52] Of the eight new breakpoints identified in this study, three (3q13.1, 3q13.2, and 9p12) do not coincide with fragile sites previously reported by these other authors. Breakpoint 3q23 identified in this study has been reported in only three people.[51,52]
Mrasek’s testing to identify fragile sites identified was performed in three Caucasian individuals (one of whom had a parent of Asian origin).[51] Liehr compared the 230 fragile sites detected by Mrasek with breakpoints found in 251 Caucasian patients with BCR and found greater than 70% overlap.[52]
In our study, we identified three breakpoints of de novo BCR not reported previously by these authors.[51,52] These three breakpoints (3q13.1, 3q13.2, and 9p12) were not reported in the large-scale studies by Warburton and Giardino,[5,7] in studies in Asia,[9–11] or in other studies with smaller sample sizes conducted in Europe, North America and Australia.[12,30–46] Rosenberg studied 1056 individuals from 52 populations and found that most genetic variation was within populations; only 3%– 5% of genetic variation was due to major group differences.[13] In 2006, Bastos-Rodrigues, using a more sophisticated methodology (a set of 40 biallelic slow-evolving short insertion-deletion polymorphisms) than the one used by Rosenberg, found greater genetic variation (12.1%) across the different population groups and confirmed Rosenberg’s identification of five well-defined groups: the Americas, Africa, East Asia, Oceania, and a cluster comprising Europe, the Middle East and Central Asia.[53] Finding these previously unreported breakpoints could be due to the unique nature of our genome with its distinctive admixture of African, European and Asian populations.
Followup Problems were reported during the perinatal period in two of the children studied, but followup studies showed that their development was completely normal. For example, the carrier of the inv(2)(p11q13) rearrangement who showed dysmorphic features at birth had subsequent normal development. Identifying dysmorphic features in the perinatal period can be difficult because sometimes the process of labor can cause physical abnormalities in the newborn that are confused with dysmorphic features.
There are some de novo BCR carriers who, even though they are apparently normal at birth, can exhibit late-onset phenotypic traits during long-term followup, primarily due to neurodevelopmental disorders, which suggest gene mutations caused by de novo chromosome rearrangement. This commonly occurs when there are cryptic chromosome aberrations.[11,30] In this study, we present the example of a woman with inv(3)(q13.1q23) who was apparently normal at birth but at the time this manuscript was being prepared had a mental disability, epilepsy, autism-like behavior, and dysmorphic features.
Two breakpoints in the long arm of chromosome 3 (3q13.1 and 3q13.2) were detected very close together. These breakpoints are not reported in the literature consulted on de novo BCR in antenatal diagnosis in the USA and Canada, Europe, Asia and Australia.[5,7,9–12,30–44] Since these breakpoints were associated with severe phenotypic abnormalities and mental disability in carrier individuals, their presence should be carefully considered during antenatal diagnosis and genetic counseling for couples. In such cases, we recommend thorough ultrasound monitoring and molecular techniques, such as FISH and microarray, to supplement diagnosis in order to detect possible cryptic genome aberrations.
Two patients with complex chromosome rearrangements involving three or more chromosomes had no neurological abnormalities. In one case, the individual had undergone antenatal diagnosis in the 1990s and was encountered again because he exhibited azoospermia and attended an infertility service. In the other case, the carrier was a completely normal 12-year-old child, for whom microarray analysis found no gain or loss of inherited material.[54] This is unusual because when several chromosomes are involved in a de novo rearrangement, the possibility of phenotype disorders increases due to poor segregation of derivative chromosomes or generation of recombinant chromosomes.[55]
One study limitation is that the genome breakpoints were detected using conventional cytogenetics, not molecular techniques such as FISH and microarray. Molecular cytogenetic analysis could have provided more sensitivity in locating breakpoints at the chromosome band and sub-band level and would have helped in comparing our breakpoints with those of Mrasek[51] and Liehr.[52] These techniques were not available to the laboratories participating in our study, in part because of when the tests were conducted and in part due to their high cost. Nevertheless, the results are relevant because the study compiled data from five Latin American laboratories and was supplemented by long-term followup of carriers of these de novo rearrangements.
Another limitation is the relatively small number of cases analyzed, compared to the Warburton and Giardino studies,[5,7] and that not all participating countries provided a sufficient number of cases. It was not our objective to make comparisons among the Latin American countries that contributed samples, as these countries also have different genetic characteristics due to diverse ancestries. We must consider that data from Mexico, Costa Rica, and Colombia only make up 6.6% of the sample.
As did previous studies, our findings suggest a relationship between constitutional breakpoints during the antenatal period and fragile sites. However, confirmation of a relationship with biological implications requires molecular analyses of breakpoints. Molecular cytogenetic techniques provide greater precision in antenatal diagnosis,[49,50] but are still unavailable in most less-developed countries. Therefore, even with the study’s limitations, it demonstrates the utility of conventional techniques for studying de novo BCRs with a focus on chromosome aberrations and their possible relationship with genome admixture in Latin American countries. Our results are useful in genetic counseling for pregnant women in Latin America.
CONCLUSIONS
Three new breakpoints were identified in the genome, related to de novo BCRs, which may be due to the typical genetic admixture in Latin America. Two of these breakpoints identified for the first time are considered high risk because they are associated with severe phenotypic abnormalities in carriers. In carriers of de novo BCRs, a minimum two-year followup period is recommended, as many phenotypic disorders have late onset.
ACKNOWLEDGMENTS
The authors thank Silvia Méndez, Luis Casaco, and Judith Pupo for their support during preparation of this manuscript. We thank all the medical geneticists and cytogenetic specialists for their considerable contribution to the study. Thank you to Joe Munch in MD Anderson’s Department of Scientific Publications for editing the manuscript.
References

- Gadner RJM, Sutherland GR, Shaffer LG. Basic concepts. In: Chromosome Abnormalities and Genetic Counseling. 4th ed. New York: Oxford University Press Inc.; 2011 Nov 11. p. 3–64.
- Bugge M, Bruun-Petersen G, Brondum-Nielsen K, Friedrich U, Hansen J, Jensen G, et al. Disease associated balanced chromosome rearrangements: a resource for large scale genotype–phenotype delineation in man. J Med Genet [Internet]. 2000 Nov [cited 2018 Jun 5];37(11):858–65. Available from: https://www
.ncbi.nlm.nih.gov/pmc/articles/PMC1734480/
- Le Scouarnec S, Gribble SM. Characterizing chromosome rearrangements: recent technical advances in molecular cytogenetics. Heredity [Internet]. 2012 [cited 2018 Jun 5];108:75–85. Available from: https://www.nature.com/articles/hdy2011100
- Feenstra I, Hanemaaijer N, Sikkema-Raddatz B, Yntema H, Dijkhuizen T, Lugtenberg D, et al. Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur J Hum Genet [Internet]. 2011 Nov [cited 2018 Jun 5];19(11):1152–60. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3198145/
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet [Internet]. 1991 Nov [cited 2018 Jun 5];49(5):995–1013. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1683246/
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340.
- Giardino D, Corti C, Ballarati L, Colombo D, Sala E, Villa N, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat Diagn [Internet]. 2009 Mar [cited 2018 Jun 5];29(3):257–65. Available from: https://obgyn.onlinelibrary.wiley.com/doi/abs/10.1002/pd.2215
- Sheth F, Rahman M, Liehr T, Desai M, Patel B, Modi C, et al. Prenatal screening of cytogenetic anomalies— a Western Indian experience. BMC Pregnancy Childbirth [Internet]. 2015 [cited 2018 Jun 5];15:90. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4396805/
- Zhang HG, Zhang XY, Zhang HY, Tian T, Xu SB, Liu RZ. Balanced reciprocal translocation at amniocentesis: cytogenetic detection and implications for genetic counseling. Genet Mol Res. 2016 Aug 19;15(3). DOI: 10.4238/gmr.15038556.
- Cheng CP, Wu CP, Lin CJ, Su YN, Chern SR, Tsai FJ, et al. Balanced reciprocal translocations detected at amniocentesis. Taiwan J Obstet Gynecol [Internet]. 2010 Dec [cited 2018 Jun 5];49(4):455–67. Available from: https://www.sciencedirect.com/science/article/pii/S1028455910600988?via%3Dihub
- Cheng CP, Chern SR, Lee CC, Lin CC, Li YC, Hsieh LJ, et al. Prenatal diagnosis of de novo t(2;18;14)(q33.1;q12.2;q31.2),dup(5)(q34q34), del(7) (p21.1p21.1), and del(10)(q25.3q25.3) and a review of the prenatally ascertained de novo apparently balanced complex and multiple chromosomal rearrangements. Prenat Diagn [Internet]. 2006 Feb [cited 2018 Jun 5];26(2):138–46. Available from: https://obgyn.onlinelibrary.wiley.com/doi/full/10.1002/pd.1369
- Sinnerbrink IB, Sherwen A, Meiser B, Halliday J, Amor DJ, Waters E, et al. Long-term health and development of children diagnosed prenatally with a de novo apparently balanced chromosomal rearrangement. Prenat Diagn [Internet]. 2013 Sep [cited 2018 Jun 6];33(9):831–8. Available from: https://obgyn.onlinelibrary.wiley.com/doi/abs/10.1002/pd.4131
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science. 2002;298(23):81–5.
- Méndez-Rosado LA, Hernández-Pérez G, Palencia-Céspedes D, Quiñones-Maza O, Barrios-Martínez A, Suárez-Mayedo U. [Structural aberration mosaicism, incidence and prenatal consequences]. Rev Cubana Genet Comunit [Internet]. 2007 [cited 2018 Jun 6];1(1):34–6. Available from: http://bvs.sld.cu/revistas/rcgc/v1n1/gco06107.pdf. Spanish.
- Quiñones-Maza OL, Méndez-Rosado LA, Quintana-Aguilar J, Suárez-Mayedo U, García-Rodríguez M, Barrios-Martínez A, et al. [Structural chromosomal rearrangement for prenatal and postnatal cytogenetic studies reference to types and carrier sex]. Rev Cubana Obstet Ginecol [Internet]. 2015 [cited 2018 Jun 6];41(1):10–3. Available from: http://bvssld.cu/revistas/gin/vol41_1_15/gin02115.htm. Spanish.
- Cerrillo-Hinojosa M, Yerena de Vega MC, González-Panzzi ME, Godoy H, Galicia J, Gutiérrez Nájar A. [Genetic amniocentesis in high-risk populations. Experience in 3081 cases]. Ginecol Obstet Mex. 2009 Apr;77(4):173–82. Spanish.
- Grether-González P, Cámara-Polanco V, Ulloa-Avilés V, Salas-Labadia C, Almanza-Márquez R, Kogan-Frenk S, et al. [Prenatal diagnosis for amniocentesis. Clinical and cytogenetic experience in 1,500 cases]. Ginecol Obstet Mex. 2010 Sep;78(9):493–503. Spanish.
- Fortes-Lima C, Bybjerg-Grauholm J, Marin-Padrón LC, Gomez-Cabezas EJ, Bækvad-Hansen M, Hansen CS, et al. Exploring Cuba–s population structure and demographic history using genome-wide data. Sci Rep [Internet]. 2018 Jul 30 [cited 2018 Aug 6];8(1):11422. Available from: http://dx.doi.org/10.1038/s41598-018-29851-3
- Mao X, Bigham AW, Mei R, Gutiérrez G, Weiss KM, Brutsaert TD, et al. A genome wide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet [Internet]. 2007 Jun [cited 2018 Jun 6];80(6):1171–8. Available from: https://ac.els-cdn.com/S0002929707610349/1-s2.0-S0002929707610349-main.pdf?_tid=5d84abe6-9fe7-4849-9cf6-a4108a8cf85e&acdnat=1528313968_95eb00e34a8b4c34a65b45cf17d3d492
- Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, et al. A genome wide single nucleotide polymorphism panel for Mexican American admixture mapping. Am J Hum Genet [Internet]. 2007 Jun [cited 2018 Jun 6];80(6):1014–23. Available from: https://ac.els-cdn.com/S0002929707610210/1-s2.0-S0002929707610210-main.pdf?_tid=a2beaa5d-4de2-4616-84c210d5ef8c6dde&acdnat=1528313865_4d868de9496423e94d2e29048cbc002c
- Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet [Internet]. 2004 May [cited 2018 Jun 6];74(5):965–78. Available from: https://ac.els-cdn.com/S0002929707643626/1-s2.0-S0002929707643626-main.pdf?_tid=e4c75ef1-002c-4fc1-a527-cf35a7736b5a&acdnat=1528313763_d3ba64a6bc1e6c63a167e8fd7982d4e7
- Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, et al. Genetic variation and population structure in Native Americans. PLoS Genet [Internet]. 2007 Nov [cited 2018 Jun 6];3(11):e185. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/pmid/18039031/
- Barch MJ, Kanutsen T, Spurbeck JL; Association of Genetic Technologists. AGT Cytogenetics Laboratory Manual. 2nd ed. New York: Raven Press; 1991. 666 p.
- Shaffer LG, Tommerup N, editors. ISCN. An International System for Human Cytogenetic Nomenclature. Basel (CH): S Karger; 2005 Dec 1. 130 p.
- Nicolaides K, Shawwa L, Brizot M. Ultrasonographically detectable markers of fetal chromosomal defects. Ultrasound Obstet Gynecol [Internet]. 1993 Jan 1 [cited 2018 Jun 6];3(1):56–9. Available from: https://obgyn.onlinelibrary.wiley.com/doi/epdf/10.1046/j.1469-0705.1993.03010056.x
- Hysert M, Bruyère H, Côté GB, Dawson AJ, Dolling JA, Fetni R, et al. Prenatal cytogenetic assessment and inv(2)(p11.2q13). Prenat Diagn [Internet]. 2006 Jul 5 [cited 2018 Jun 7];26(9):810–13. Available from: https://obgyn.onlinelibrary.wiley.com/doi/epdf/10.1002/pd.1508
- Fickelscher I, Liehr T, Watts K, Bryant V, Barber JCK, Heidemann S, et al. The variant inv(2)(p11.2q13) is a genuinely recurrent rearrangement but displays some breakpoint heterogeneity. Am J Hum Genet [Internet]. 2007 Oct [cited 2018 Jun 7];81(4):847–56. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2227935/pdf/AJHGv81p847
- Stoll C, Flori E, Macler J, Renaud R. Prenatal diagnosis and postnatal follow-up of an abnormal child with two de novo apparently balanced translocations. Hum Genet. 1979 Mar;47(2):221–4.
- Bogart MH, Bradshaw CL, Jones OW, Schanberger JE. Prenatal diagnosis and follow-up of a child with a complex chromosome rearrangement. J Med Genet [Internet]. 1986 Apr [cited 2018 Jun 7];23(2):180–3. Available from: http://jmg.bmj.com/content/jmedgenet/23/2/180.full.pdf
- Kim HJ, Perle MA, Bogosian V, Greco A. Prenatal diagnosis of a de novo complex chromosomal rearrangement involving four chromosomes. Prenat Diagn. 1986 May–Jun;6(3):211–6.
- Köhler J, Brackertz M, Feige A, Grimm T. Prenatal diagnosis of a complex, balanced rearrangement of chromosomes 7 and 14 in a healthy child with a history of preconceptual X-ray exposure. Prenat Diagn. 1986 Sep–Oct;6(5):389–90.
- Pruggmayer M, Zoll B, Leipoldt M, Thies U. Prenatal diagnosis and postnatal follow-up of a child with two de novo unrelated balanced reciprocal translocations. Prenat Diagn. 1990 May;10(5):337–42.
- Iqbal M, Ramadan S, Ali F, Kurdi W. Complex de novo cryptic subtelomeric rearrangements in a fetus with multiple ultrasonographic abnormalities and a normal karyotype at amniocentesis. Prenat Diagn. 2005;25:1142–9.
- Batista DAS, Tuck-Muller CM, Martínez JE, Kearns WG, Pearson PL, Stetten G. A complex chromosomal rearrangement detected prenatally and studied by fluorescence in situ hybridization. Hum Genet. 1993 Sep;92(2):117–21.
- Sikkema-Raddatz B, Sijmons RH, Tan-Sindhunata MB, van der Veen AY, Brunsting R, de Vries B, et al. Prenatal diagnosis in two cases of de novo complex balanced chromosomal rearrangements. Three-year follow-up in one case. Prenat Diagn. 1995 May;15(5):467–73.
- Cotter PD, Caggana M, Willner JP, Babu A, Desnick RJ. Prenatal diagnosis of a fetus with two balanced de novo chromosome rearrangements. Am J Med Genet. 1996 Dec 11;66(2):197–9.
- Mercier S, Fellmann F, Cattin J, Bresson JL. Molecular analysis by fluorescence in situ hybridization of a prenatally detected de novo complex chromosomal rearrangement t(2q;3p;4q;13q). Prenat Diagn. 1996 Nov;16(11):1046–50.
- Ruiz C, Grubs RE, Jewett T, Cox-Jones K, Abruzzese E, Pettenati MJ, et al. Prenatally diagnosed de novo apparently balanced complex chromosome rearrangements: two new cases and review of the literature. Am J Med Genet. 1996 Aug 23;64(3):478–84.
- Calabrese G, Morizio E, Franchi PG. De novo complex chromosome rearrangement detected by fluorescence in situ hybridization on amniotic fluid cells. Am J Med Genet. 1998 Feb 3;75(4):414–5.
- Phelan MC, Blackburn W, Rogers RC, Crawford EC, Cooley NR Jr, Schröck E, et al. FISH analysis of a complex chromosome rearrangement involving nine breakpoints on chromosomes 6, 12, 14 and 16. Prenat Diagn. 1998 May 4;18(11):1174–80.
- Hastings RJ, Watson SG, Chitty LS. Prenatal findings of a fetus with mosaicism for two balanced de novo chromosome rearrangements. Prenat Diagn. 1999 Jan;19(1):77–80.
- Peschka B, Leygraaf J, Hansmann D, Hansmann M, Schröck E, Ried T, et al. Analysis of a de novo complex chromosome rearrangement involving chromosomes 4, 11, 12 and 13 and eight breakpoints by conventional cytogenetic, fluorescence in situ hybridization and spectral karyotyping. Prenat Diagn. 1999 Dec;19(12):1143–9.
- Balícek P, Jüttnerová V, Jarosová M, Fialová J, Fiedler Z, Kolmanová J. [Prenatal diagnosis of de novo complex balanced chromosome rearrangements involving in chromosomes 3, 4, and 13]. Cas Lek Cesk. 2001 Mar 1;140(4):122–4. Czech.
- Vasilevska M, Ivanoska E, Kubelka K, Sukarova-Angelovska E, Dimeska G. The incidence and type of chromosomal translocations from prenatal diagnosis of 3800 patients in the Republic of Macedonia. Balkan J Med Genet [Internet]. 2013 Dec [cited 2018 2018 Jun 7];16(2):23–8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4001411/pdf/bjmg-16-02-23.pdf
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002 Feb;18(2):74–82.
- Kato T, Kurahashi H, Emanuel BS. Chromosomal translocations and palindromic AT-rich repeats. Curr Opin Genet Dev [Internet]. 2012 Jun [cited 2018 Jun 7];22(3):221–8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3378763/pdf/nihms362914.pdf
- Schluth-Bolard C, Delobel B, Sanlaville D, Boute O, Cuisset JM, Sukno S, et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet. 2009 Sep–Oct;52(5):291–6.
- Feuk L. Inversion variants in the human genome: role in disease and genome architecture. Genome Med [Internet]. 2010 Feb 12 [cited 2018 Jun 7];2(2):11. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2847702/pdf/gm132.pdf
- Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet [Internet]. 2017 Jan [cited 2018 Jun 7];49(1):36–45. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5307971/
- Ordulu Z, Kammin T, Brand H, Pillalamarri V, Redin CE, Collins RL, et al. Structural chromosomal rearrangements require nucleotide-level resolution: lessons from next-generation sequencing in prenatal diagnosis. Am J Hum Genet [Internet]. 2016 Nov 3 [cited 2018 Jun 7];99(5):1015–33. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5097935/pdf/main.pdf
- Mrasek K, Schoder C, Teichmann AC, Behr K, Franze B, Wilhelm K, et al. Global screening and extended nomenclature for 230 aphidicolin inducible fragile sites, including 61 yet unreported ones. Int J Oncol. 2010 Apr;36(4):929–40.
- Liehr T, Kosayakova N, Schröder J, Ziegler M, Kreskowski K, Pohle B, et al. Evidence for correlation of fragile sites and chromosomal breakpoints in carriers of constitutional balanced chromosomal. Balkan J Med Genet [Internet]. 2011 [cited 2018 Jun 7];14(2):13–6. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3776699/pdf/bjmg-14-02-13.pdf
- Bastos-Rodrigues L, Pimenta JR, Pena SDJ. The genetic structure of human populations studied through short insertion-deletion polymorphisms. Ann Human Genet [Internet]. 2006 [cited 2018 Jun 7];70:658–65. Available from: https://onlinelibrary.wiley.com/doi/epdf/10.1111/j.1469-1809.2006.00287.x
- Quadrelli A, Vaglio A, Quadrelli R, Mechoso B, Fan YS, Huang T. High density array comparative genomic hybridization analysis and follow-up of a child with de novo complex chromosome rearrangement detected prenatally. Prenat Diagn [Internet]. 2007 Oct [cited 2018 Jun 7];27(10):982–3. Available from: https://obgyn.onlinelibrary.wiley.com/doi/epdf/10.1002/pd.1831
- Gadner RJM, Sutherland GR, Shaffer LG. Parent with chromosomal abnormality. In: Chromosome Abnormalities and Genetic Counseling. 4th ed. New York: Oxford University Press Inc.; 2011. p. 212–20.
THE AUTHORS
Luis A. Méndez-Rosado (Corresponding author: albermen@infomed.sld.cu), biologist with a master’s degree in medical genetics and doctorate in health sciences, Cytogenetics Laboratory, National Medical Genetics Center (CNGM), Havana, Cuba.
Olga Quiñones-Maza, biologist with a master’s degree in medical genetics, CNGM, Havana, Cuba.
Alicia Vaglio, physician specializing in genetics, Hospital of Montevideo, Uruguay.
Roberto Quadrelli, physician specializing in genetics, Hospital of Montevideo, Uruguay.
Diana Sánchez-Peñarate, physician specializing in clinical genetics, Biogenetics Diagnosis SAS, Bogotá, Colombia.
Michel Soriano-Torres, microbiologist with a master’s degree in bioinformatics, Cytogenetics Laboratory, CNGM, Havana, Cuba.
Mabel Cerrillo-Hinojosa, physician specializing in clinical genetics, AGN Reproduction and Genetics Group (AGN) and 20 de Noviembre National Medical Center (CMN) Institute for Social Security and Services for State Workers (ISSSTE), Mexico DF, Mexico.
Fernando Escobedo-Aguirre, physician specializing in clinical genetics, AGN and 20 de Noviembre CMN, ISSSTE Mexico DF, Mexico.
Alfonso Gutiérrez-Nájar, physician specializing in clinical genetics, AGN and 20 de Noviembre CMN, ISSSTE Mexico DF, Mexico.
Patricia Venegas-Barbosa, physician, National Children’s Hospital, San José, Costa Rica.
Anduriña Barrios-Martinez, biologist with a master’s degree in medical genetics, CNGM, Havana, Cuba.
Dulce Hechavarría-Estenoz, physician specializing in clinical genetics, Cytogenetics Laboratory, Provincial Medical Genetics Center (CPGM), Santiago, Cuba.
Pedro Carbonell-de la Torre, biologist with a master’s degree in medical genetics, CPGM, Sancti Spíritus, Cuba.
Héctor I. Pimentel-Benitez, biologist with a master’s degree in medical genetics, Cytogenetic Laboratory, CPGM, Camagüey, Cuba.
Orlando González-Salé, cytogeneticist, CPGM, Camagüey, Cuba.
José Hernández-Gil, agronomist with a master’s degree in medical genetics, Cytogenetics Laboratory, CPGM, Holguín, Cuba.
María Elena de la Torre-Santos, physician specializing in clinical genetics, Cytogenetics Laboratory, CPGM, Villa Clara, Cuba.
Yamile Alonso-García, biologist, Cytogenetics Laboratory, CPGM, Guantánamo, Cuba.
Niurka Cedeño-Aparicio, biologist, CPGM, Granma, Cuba.
Patricia Torriani-Mendoza, biologist, Cytogenetics Laboratory, CNGM, Havana, Cuba.
Enny Morales-Rodríguez, biologist with a master’s degree in medical genetics and doctorate in health sciences, Cytogenetics Laboratory, CNGM, Havana, Cuba.
Diana Martín-García, physician specializing in clinical genetics with a doctorate in medical sciences, CPGM, Sancti Spiritus, Cuba.
Elizabeth Cuétara-Lugo, biochemist with a doctorate in biological sciences, Comandante Manuel Fajardo Medical Faculty, Medical University of Havana, Cuba.
Niurka González-Domínguez, physician specializing in clinical genetics, Ramón González Coro Maternity Hospital, Havana, Cuba.
Peter Hu, molecular geneticist. Director, Diagnostic Molecular Science Programs,
School of Health Professions, University of Texas MD Anderson Cancer Center, Houston, Texas.
Submitted: May 07, 2018 Approved: October 07, 2018 Disclosures: None









