INTRODUCTION
Since the description in 1984 of the first person infected with human immunodeficiency virus type 1 (HIV-1), the pandemic has affected more than 150 countries. UNAIDS reported 34.2 million people living with HIV worldwide in 2013.[1]
The substantial efforts made over the past three decades to develop an effective vaccine against HIV have not yet yielded success, and scientific, social and ethical difficulties remain major challenges for medical researchers. However, combined treatment with four families of antiretrovirals—inhibiting HIV replication by acting on different targets of its mechanism—have enabled HIV infection to become a chronic disease with acceptable quality of life.[2]
HIV, a lentivirus belonging to the Retroviridae family, is a complex retrovirus differing from other viruses by its extraordinary variability, expressed in groups, subtypes, sub-subtypes, circulating recombinant forms (CRF) and unique recombinant forms (URF). Despite its great variability, phylogenetic analysis of numerous HIV-1 isolates from diverse geographic origins has enabled its classification in four distinct phylogenetic groups: M, O, N and
P. The M group includes most of the variants responsible for the global pandemic: subtypes A, B, C, D, F, G, H, J and K, and sub-subtypes A1, A2, A3, A4, A5, F1 and F2. To date, 61 CRFs have been described, as well as multiple URFs.[3]
HIV diversity is the combined result of various forces acting separately: a) frequent introduction of mutations by reverse transcriptase (because of errors and lack of corrective action); b) high replication rate or rapid virus turnover in vivo; c) selective immune response pressure; d) therapeutic pressure and e) genetic recombination as part of the retrovirus replication mechanism.[4,5]
HIV genetic diversity should be studied not only to discern its origin and understand its molecular epidemiology, but to also monitor emergence of new variants that may be more transmissible or pathogenic, as well as their implications for serological and molecular laboratory diagnosis, changes in resistance patterns to antiretroviral drugs, and development of an effective vaccine.[5]
Early HIV-1 genetic characterization in Cuba demonstrated circulation of several viral variants: subtypes A, B, C, D, F, G and H of the M group. Subsequently, a gradual increase in diversity has been observed, with emergence of new subtypes and recombinant forms caused by a mixture of subtypes that originally contributed to the epidemic in Cuba.[6–14]
Kanki demonstrated presence of CRFs in 1997, in samples from serum banks collected in the 1980s (when molecular findings considered only pure subtypes).[15] This suggests that recombination is not a recent phenomenon, but goes back to the 1980s, when nucleic acid sequencing technology for complete genome studies was not available. It was not until the late 1990s that circulation of multiple subtypes and their recombinations was reported.[2,5]
The same may have occurred in Cuba, and diversity has certainly increased. Early studies targeted the envelope (env) gene, since it is the most important for immune response and vaccine studies; subsequently, the gag and pol genes were studied, revealing circulation of other subtypes and CRFs. It was not until 2001 that discussion of HIV mosaicism in Cuba began.[9] Later research found various complex forms and a CRF increase in the seropositive population.[10–14]
While interest has grown worldwide in the association of HIV-1 genetic variability with transmissibility and clinical disease progression, few studies have successfully compared subtypes, because the necessary clinical and epidemiologic information about patients and their contacts is not readily available. Factors such as stigma and discrimination can introduce bias in available information; patients, for example, may be reticent to report multiple sexual partners, making it difficult to trace contacts for epidemiologic monitoring.[5,6]
Serological and molecular studies in Cuba show low prevalence of seropositivity, but increasing genetic variability with epidemiologic implications.[6–14] Our objectives, therefore, were to study the genetic variability of the HIV-1 env, gag and pol structural genes in Cuba; determine the prevalence of B and non-B subtypes according to epidemiologic and behavioral variables; and establish whether relationships exist between genetic variability and transmission, and between such variability and clinical disease progression in persons living with HIV/AIDS in Cuba.
METHODS
Design and Population This is a descriptive study, the universe being the 17,625 HIV-1 seropositive people in Cuba between January 1, 2008, and December 31, 2012. The study population comprised 590 persons (480 men and 110 women) aged 14–70 years, or 3.3% of all seropositive individuals in Cuba. Selection was made by nonrandom sampling, proportional to HIV prevalence by province, including samples that met inclusion criteria until the assigned quota was filled. Table 1 shows the universe and study population by province and totals.[1,16]
Table 1: Universe and study population distribution by province
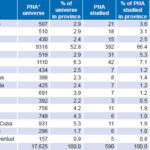
Source: National Reference Laboratory/National STI–HIV/AIDS Program
* PHA: person living with HIV/AIDS
Inclusion criteria HIV-1 seropositive individuals with infection confirmed by serological methods, of both sexes, from each of Cuba’s 15 provinces and the Isle of Youth Special Municipality, who voluntarily agreed to participate and provide the necessary epidemiologic information were eligible.
Ethics Procedures were carried out according to the ethical standards of the Ministry of Public Health (MINSAP) and the Ministry of Science, Technology and Environment, in accordance with the principles of the Declaration of Helsinki. The study protocol was approved by the ethics committee of the National AIDS Reference Laboratory (NRL) of the National STI–HIV/AIDS program. At the time of sampling for the molecular study, each patient provided written informed consent and filled out an epidemiologic questionnaire. Procedures for handling patient information ensured confidentiality of individual identities.
Sampling and nucleic acid extraction A 10 mL peripheral blood extraction was performed by venipuncture with anticoagulant (EDTA 0.2 mmol/L, pH 7.2). Samples were processed within 24 hours of extraction. To obtain the genetic material two procedures were used: the first method for obtaining DNA by Ficoll gradient (GE Healthcare, Sweden), phenol-chloroformisoamyl alcohol (Sigma-Aldrich, USA) extraction and precipitation in absolute alcohol (Merck KGaA, Germany);[17,18] the second procedure for obtaining DNA or RNA, using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Germany), according to manufacturer’s instructions. The first procedure was used for all samples characterized by env/gag heteroduplex mobility assay, and the second for all samples characterized by automated nucleic acid sequencing of the env/polgenes.
Molecular studies Two methods were employed for molecular genotyping. From 2008 through 2010 the heteroduplex mobility assay (HMA) was used for genes env and gag using the HMA sub-typing kit (NIH AIDS Research and Reference Reagent Program, USA).[19,20]. Proviral DNA was amplified by nested PCR for the gag and env genes. For the envelope gene, ED5/ED12 and ES7/ES8 or ED31/ED33 primers were used that recognize a fragment of the env gene encoding the V1–V5 region of gp120.[20] For the gag gene, H1G777/H1P202 and H1Gag1584/g17 primers were used that recognize a fragment of the gag gene.[21] The heteroduplexes were formed between the samples and reference standards. Recognition between the two DNA fragments generated heteroduplex formations, which were run on polyacrylamide gel. Their relative mobility on the gel reflected their homology.[20–21] Genetic distance between sequences and subtypes for the env/gag genes was determined.
In 2011 and 2012 automated nucleic acid sequencing of the env and pol genes was used, according to procedures described by Thomson[22] and Quarleri.[23]
Plasma viral load was determined for each sample to define RNA copies/mL in plasma as the criterion for genetic material amplification. The COBAS AmpliPrep/COBAS TaqMan HIV-1 Test (Roche Diagnostics GmbH, Germany) was used, following manufacturer’s specifications. Proviral DNA was amplified by nested PCR for the env gene (C2-V3-C3 fragment) using external primers ED5/ED12 and internal primers ED31/ED33.[20] For the pol gene, primers for protease and reverse transcriptase fragments were used: the pair RT3.1/5’CP as external primers and three sets (1F/6B, A35/ NE1 and RT3208F/RT3798R) as internal primers.[23] The purified PCR product was directly sequenced with the QuickStart kit (Beckman Coulter, USA), according to manufacturer’s instructions, in a CEQ 8800 automated sequencer (Beckman Coulter, USA).
Phylogenetic analysis The resulting pol and env gene sequences were assembled and edited using the Sequencher 4.0.5 program (Gene Codes Corp, USA). Multiple sequence alignment was done with the Muscle program (MEGA 6.0, USA). A basic local alignment search tool was applied to compare results with subtype reference sequences from the Los Alamos National Laboratory database.[24] Phylogenetic analysis was performed with MEGA version 6.0.[25] The phylogenetic tree was made using the neighbor-joining method and genetic distance was calculated according to Kimura’s two parameters.[26] Bootstrapping values were calculated based on 1000 replications. Analysis of recombinant inter-subtypes and recombination hotspots was performed by bootscanning using SimPlot version 3.5.1.[26]
Variables The main variables were subtype (A, B, C, D, F, G, H, J and K), sub-subtype (A1, A2, A3, A4, A5 F1 and F2), CRF and URF.[24]
Distribution of secondary variables by sex, sexual preference (men who have sex with men: MSM; heterosexual: HT), place of infection and place of residence (Cuba, province, region) was analyzed. Disease transmission, clinical progression (mean survival time in years and average AIDS-free time in years, assessed by CD4 count and plasma viral load, data from SIDATRAT, Pedro Kourí Tropical Medicine Institute) were analyzed by subtype (B, non-B).[27]
For molecular epidemiology analysis, genotyping and patients´ clinical–epidemiologic identification data were considered: year of diagnosis, age, sex (male, female), env gene subtype, gag gene subtype, pol gene subtype, death from AIDS, province, probable date and place of infection, date of diagnosis, date classified as AIDS, date of death, and sexual orientation (HT, MSM). Data were taken from the PACIENTE database of MINSAP’s National Epidemiology Program (PNE).
Geographic distribution analysis was performed at national, provincial and regional levels. Provinces were grouped in three regions: West (Pinar del Río, Mayabeque, Artemisa, Havana, Matanzas and Isle of Youth), Center (Villa Clara, Cienfuegos, Sancti Spíritus, Ciego de Ávila and Camagüey) and East (Las Tunas, Holguín, Granma, Santiago de Cuba and Guantánamo).
To associate variability with transmissibility of B and non-B subtypes, three variables were analyzed: reported contacts, studied contacts and positive contacts (data from CONTACTOS database, PNE/MINSAP).
To associate clinical disease progression in B and non-B subtypes, four time lapses (in years) were studied: from probable date of infection (PDI) to date patient was diagnosed with AIDS (DxAIDS); from date of HIV diagnosis (DxHIV) to DxAIDS; from PDI or DxHIV to date of death; and from DxAIDS to date of death (data from PACIENTE database, PNE/MINSAP).
Database variables that complement analysis of clinical disease progression were also taken into consideration: namely, persons who developed the disease (AIDS), persons who died, living persons, classification of patients into fast progressors (FP), typical progressors (TP), and slow progressors (SP). The CD4+ cell count, in percent and absolute value, was used for classifying patients as FP, when the CD4+ cell count was < 300 between 1 and 3 years; TP, from 4 to 9 years and SP, ≥10 years. Another variable analyzed was viral load (VL), taking as baseline values ≥1000 RNA copies/mL or 3.0 log in plasma between 1 and 3 years, 4 and 9 years and ≥10 years, respectively,[28] (data from SIDATRAT).[27]
In the group of patients classified as FP, it was necessary to consider whether other epidemiologic and behavioral conditions favoring transmission and clinical progression of the disease were involved. The conditions studied were >5 lifetime sexual partners reported, intravenous drug use, ongoing sexual relationships with seropositive partners or contacts, age >40 years at the time of primary infection (Database PACIENTE, PNE/MINSAP).
Analysis Parametric and nonparametric methods and the Z and chi square statistics were used to compare means and proportions, respectively. Actuarial tables were prepared for analyzing clinical progression and survival times using Kaplan–Meier analysis and the log rank test. Statistica 8.0 (StatSoft, USA) was used for data processing and EPIDAT 3.1 (Spain) for epidemiologic analysis of tabulated data.[16,29] The statistical significance level for all tests was p < 0.05. Comparisons were made between B and non-B subtypes for each of the secondary variables described above.
RESULTS
Out of 17,625 people seropositive for HIV-1 in the study period, isolates from 590 were studied. Of them, 566 (96%) could be typed for two or three structural genes env/gag or env/pol, by any one of the methods used for characterization: 297 (50.3%) subtype B and 269 (45.5%) non-B subtype; 24 (4.1%) were not typeable (Table 2).
Molecular findings and clinical-epidemiologic data The relationship between B and non-B subtypes with sex and sexual orientation was as follows: subtype B was more frequent in men (252/456; 55.3%) than in women (45/110; 40.9%), a highly significant difference (p < 0.001). Similarly, analysis of sexual orientation found subtype B more frequent in MSM (232/401; 57.9%) than in HT (65/165; 39.4%), a highly significant association (p <0.001).
Persons were infected with HIV-1 in 12 countries; mostly in Cuba (subtype B: 277; 93.3% and non-B: 249; 92.6%). Infections occurred in six African countries (Republic of Congo, Ethiopia, South Africa, Zambia, Guinea, Angola), and the USA, Canada, Spain, Costa Rica and Brazil. Five persons, all sailors, did not report the place of infection.
Table 2: HIV-1 subtypes and recombinant forms of env/gag/pol genes (n = 590)

CRF: circulating recombinant forms
URF: unique recombinant forms
Subtypes observed in the study and introduced in Cuba were: A, B, C, D, F, G, H, ADK, A/AG, CRF02 and CRF18. Distribution of B and non-B subtypes in Cuba is shown in Table 3. Both B and non-B subtypes were found throughout the country. Some differences between provincial proportions of B and non-B subtypes were found, but there were no significant differences by region (Table 3).
Analysis of B and non-B subtype transmission is shown in Table 4. Persons with subtype B reported a larger number of contacts, of which 75.2% could be studied to confirm the serologic diagnosis of HIV-1. For those with non-B subtypes, 90.5% of contacts reported could be studied. There was no significant difference between B and non-B subtypes in the ratio of positive contacts to studied contacts (p = 0.269). A similar analysis was performed for non-B subtypes and the CRFs most frequently found in the studied sample, again, with no significant differences detected.
Table 5 displays analysis of clinical disease progression for B and non-B subtypes according to whether or not the person had developed AIDS at the time of analysis. There were no significant differences between the two groups. Another analysis of disease progression was performed for each group considering time elapsed, in years, from PDI to DxAIDS. The difference between the two groups was not significant. The same analysis was also performed for PDI to DxHIV: the average time between PDI and DxHIV of all patients included in the study was estimated at 2.4 years. Analysis of time in years from DxHIV to DxAIDS yielded similar results, with a nonsignificant difference of less than one year between persons with subtype B and those with non-B.
In terms of disease progression (Table 5), a higher proportion of persons infected with subtype B died during the study period than did persons with non-B subtypes (25.2% vs. 12.6%, respectively). This difference was significant. No statistically significant difference was found in survival times between persons with B and non-B subtypes. Regarding time from DxAIDS to death, lifespan of persons with subtype B was one year shorter than for those with non-B subtypes, but the difference was not statistically significant.
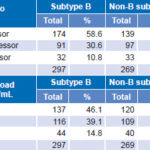
Regarding rate of progression as measured by CD4 count, no statistically significant differences were found between B and non-B subtypes, nor were differences observed between the two groups when we analyzed time elapsed to reach a viral load of >1000 copies/mL (Table 6).
When epidemiologic and behavioral variables were analyzed in the FP patient group (174 with subtype B and 139 non-B), several fac tors were frequently observed in the same patient. No significant differences were found between patients with B and non-B subtypes with respect to having one or more risk factors, although some differences were observed for individual factors (Table 7).
Among these, the one displaying the greatest difference is more than five sexual partners. These differences were significant for subtype B compared with non-B. Subtype B in this study was associated with the highest risk group, men with the sexual orientation MSM. In many patients, having >5 sexual partners coexisted with other risk factors, such as IV drug use, which was present in 21.4% (67/313), with no difference between subtypes (overall prevalence of IV drug use in the sample was 13.4%; 79/590). Patients with non-B subtypes tended to have stable relationships with seropositive sexual partners, although the difference did not reach statistical significance (Table 7).
Table 3: Distribution of B and non-B HIV-1 subtypes by province and region
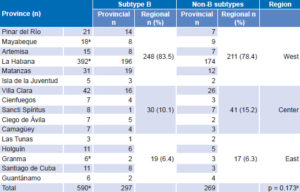
a24 not typeable: 1 in Mayabeque; 22 in La Habana; 1 in Granma
bchi square
Table 4: Transmission of B and non-B subtypes, contact tracing (n = 566)
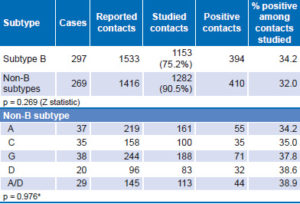
*chi square
Table 5: Clinical progression (AIDS-free time) for B and non-B subtypes (n = 566)
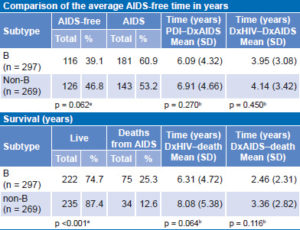
DxAIDS: date of AIDS diagnosis DxHIV: date of HIV diagnosis
PDI: probable date of infection
achi squar
bKaplan–Meier survival analysis, log-rank test
Table 6: Clinical progression by subtype, by CD4 count and plasma viral load levels (n = 566)
*chi square
Table 7: Risk factors in fast progressors with B and non-B subtypes (n = 313)

All patients aged >40 years at time of infection showed rapid progression of the disease; 22.4% of fast progressors with subtype B and 31.7% of fast progressors with subtype non-B were aged >40 years at time of infection; the difference between subtypes was not statistically significant (Table 7).
DISCUSSION
As of December 2012, Cuba’s National HIV/AIDS Program had recorded a total of 19,781 persons diagnosed HIV-1 seropositive since 1986. Although low prevalence of HIV infection in Cuba (0.2%) ranks our epidemic as one of the smallest in the world,[1] it shows high genetic diversity of subtypes, CRFs and UFRs, since favorable conditions for coinfections and superinfections have led to the recombinations between subtypes observed during our epidemiologic and molecular history.[6–14]
This study, using two nucleic acid technologies, env/gag HMA and nucleic acid sequencing for three of the HIV-1 structural genes, corroborated the aforementioned and found predominantly subtype B, but also showed that virtually all non-B subtypes were in circulation, as well as many recombinant variants corresponding to subtypes reported in previous studies. These results relate to the origins of HIV-1 variants introduced in Cuba: the coexistence of persons living with HIV/AIDS for several years in sanatoriums, the fact that several routes of infection have coincided, and epidemiologic and behavioral risk factors in the seropositive population—all contributing to the high genetic variability of HIV-1 observed in Cuba.[9,10]
Subtype B was found more frequently than any of the non-B subtypes. Early in the HIV epidemic, subtype B—which prevailed in Eastern European countries and the USA—was referred to as the predominant subtype globally, possibly because most molecular studies were carried out in those developed countries.[30] But worldwide distribution of HIV-1 viral variants has changed. Today subtype B is found in only 10% of new HIV infections globally; non-B subtypes and CRFs[31–35] account for the remaining 90%. In Cuba, however, subtype B accounts for most infections in the seropositive population, as has been consistently shown from the first molecular characterization studies. This can be explained by the fact that subtype B had its founder effect in MSM, the population at highest epidemiologic risk and the majority in our epidemic, although a gradual increase of non-B subtypes and CRFs has been observed in recent years.[12–14]
Male sex predominated in both B and non-B subtype groups because the AIDS epidemic in Cuba has occurred mostly among men (80%), and of these, 89.0% are MSM.[36] Subtype B in Cuba is associated with male sex and MSM, whereas non-B subtypes are associated with women and heterosexual orientation.
At the beginning of the epidemic in Cuba in the 1980s, subtype B became established among men, mainly MSM.[7] Non-B subtypes, on the other hand, had their founder effect in heterosexual persons. Different viral variants were introduced into Cuba by persons (mainly heterosexual men) who acquired the infection in various African countries and upon their return passed it on to female sexual partners, accounting for the HT sexual orientation predominance.[7,8] Women are more vulnerable to acquiring both B and non-B subtypes, because of the high proportion of seropositive men with bisexual practices.[1,27] These epidemiologic conditions still describe our epidemic after more than 20 years, with predominance of subtype B relative to non-B subtypes with their multiple recombinants.
Our research also delved into the molecular characteristics of the Cuban epidemic, since 93% of samples were from people infected in Cuba. Nevertheless, variants introduced from several countries were found, providing evidence of subtypes introduced into Cuba from Africa, Europe, and North and South America.[6–14] It was also observed that B and non-B subtypes, as well as CRFs, are distributed throughout the country.
Transmission analysis showed that B and non-B subtypes have been transmitted in equal proportions in the seropositive population under study. According to Zhang in 2007, HIV-1 transmission may depend on the genetic form or the presence of recombination because of changes in the properties of the viruses involved and the biological and genetic characteristics of both viruses. In this study, however, no association between genetic subtypes and transmissibility was found.[37] Transmissibility may also depend on other factors, such as levels of viral load in plasma, since high levels of viral load in HIV promote rapid progression to AIDS and increase transmissibility to subsequent contacts.[28] Jennes suggests that certain subtypes, such as subtype C, are transmitted more efficiently by perinatal route than subtypes B, A and D.[33] Kouyos in 2010, observed that non-B subtypes (A, C, D) were adapted to heterosexual transmission, while B subtype showed transmission efficiency among MSM and IV drug users.[32,38] These results are consistent with our that subtype B predominates in men and among these in MSM and non-B, in women with heterosexual orientation.
While there are no published data on Cuban prevalence of IV drug use in Cuba, the 13.4% overall prevalence and 21.4% among fast progressors we found is undoubtedly much higher than in the general population.
Study of disease progression among those infected with B and non-B subtypes showed no differences after analyzing the state of the immune system (CD4 cells) of patients classified as FP, TP and SP. Moreover, the level of plasma viral RNA or viral load, which reflects degree of virus replication, also showed no subtype-related differences in disease progression. These parameters are related because decrease in CD4 cell count is determined by elevation of viral RNA concentration in plasma.[28,34,39]
The results of progression analysis further showed that the proportion of deaths was higher in subtype B patients. Although times from probable infection date to diagnosis, from diagnosis to AIDS development and to death were shorter in subtype B patients, these differences did not reach statistical significance. The proportion of patients who developed AIDS was not significantly different among subtypes either. At the same time, subtype B was significantly associated with other risk factors such as male sex, MSM sexual orientation, and many sexual partners. It is possible that these factors played a role in the higher proportion of deaths among subtype B-infected patients, since there is no evidence to suggest that subtype B is more lethal than others. This aspect requires further research, including multivariate statistical analysis.
These observations are consistent with those made by authors who suggest that disease transmission and progression depend on many factors, such as host susceptibility (chemokine coreceptor polymorphism),[40] genetics (HLA system genotypes), the immune system (direct HIV immune response),[41] age of patient at time of seroconversion,[42] coinfections with other HIV variants or other viral agents such as hepatitis B and C viruses,[43,44] HIV genetic variability,[45,46] characteristics of the viral strain (deletions in the nef gene),[40] and its replication ability,[47] all of which could influence rapidity of AIDS progression. That is, both genetic and viral factors (of which viral subtype is only one) can be influential.
CONCLUSION
This study furthered understanding of HIV molecular epidemiology in Cuba and provided the National STI–HIV/AIDS Program with information on circulating viral variants and their behavior in the seropositive population, important for managing the epidemic, which shows high variability. The study also demonstrated subtypes and recombinant forms introduced into Cuba from several countries in a high percentage of the Cuban seropositive population and both B and non-B subtypes and recombinant forms found throughout the country. Findings suggest that HIV-1 B and non-B subtypes in Cuba do not differ in transmissibility; the question of clinical disease progression by subtype requires further research. Although HIV-1 prevalence in Cuba is low, its high genetic diversity creates a complex scenario for setting national strategies, surveillance of antiretroviral drug resistance, and use of future vaccines.










