INTRODUCTION
In the Caribbean region, acute diarrheal diseases (ADD) caused by the Vibrio genus have increased in recent years, following the earthquake in Haiti in 2010, after which WHO recommended strengthening epidemiological surveillance and national preparedness in neighboring countries to detect outbreaks immediately.[1] In June 2012, the Cuban Ministry of Public Health (MINSAP) reported a cholera outbreak in Granma Province, with 417 confirmed cases and 3 deaths. In August 2013, a new outbreak occurred in the provinces of Havana, Santiago and Camagüey, with more than 163 cases reported to PAHO.[2] In 2015, 386,507 ADD cases were reported, of which 65 were confirmed to be cholera, while in 2016, 288,832 cases of ADD and no cholera cases were reported.[3,4]
Vibrio cholerae is classified by its capsular lipopolysaccharide in more than 200 serogroups, divided into O1, O139 and non-O1, non-O139, of which only O1 and O139 are epidemic. V. cholerae O1 is biochemically identical to non-O1, non-O139 V. cholerae.[5,6] The latter are isolated in patients with clinical manifestations ranging from mild diarrhea to severe dehydration, but do not present epidemic potential. In addition, they are associated with extraintestinal symptoms.[5,7] In Cuba, the circulation of non-O1, non-O139 V. cholerae has been reported in outbreaks and sporadic cases.[8,9]
The pathogenic mechanisms involved in diarrhea caused by these enteropathogens are still not well established; a different pathogenesis is proposed for nonepidemic serogroups, in which heterogeneous virulence factors intervene.[8,10,11] In Latin America and the Caribbean, there are few studies on virulence factors in isolates of non-O1, non-O139 V. cholerae from patients with ADD.[8] Extracellular products are described, such as a thermostable enterotoxin NAG-ST, pili coregulating toxin and the production of extracellular enzymes such as gelatinase, elastase and lecithinase.[11] Diarrhea caused by V. cholerae can be treated with hydration, and antimicrobials, if necessary. To date there are no vaccines for these serogroups.[12]
The emergence of antimicrobial resistance is a global phenomenon. International studies have described V. cholerae isolates resistant to multiple antimicrobials, with increased resistance to furazolidone, nalidixic acid, sulfisoxazole, streptomycin and trimethoprim–sulfamethoxazole, and decreased sensitivity to ciprofloxacin.[1,12] Multidrug resistance (MDR) is common in strains of V. cholerae isolated from patients with intestinal and extraintestinal infections, so carrying out antimicrobial susceptibility studies is necessary and useful for establishing successful therapies.[13,14]
Determination of V. cholerae antimicrobial susceptibility is not serogroup specific. Cutoff points established by the Clinical Laboratory Standards Institute (CLSI) are determined for the species, regardless of serogroups, since intrinsic resistance to antimicrobials is associated with acquisition of genetic determinants of resistance and mutations, within species and among enteropathogens in general.[14,15]
Some antimicrobial susceptibility studies of V. cholerae have been done in Cuba. In 2004, non-O1, non-O139 V. cholerae isolates from several Cuban provinces were obtained, showing resistance to ampicillin (14%) and trimethoprim–sulfamethoxazole (16%), while 98% showed sensitivity to tetracycline and 96% to chloramphenicol.[16] A 2008 study found 32.3% and 30.7% of isolates resistant to sulfonamide and ampicillin, respectively, and percentages of sensitivity against the antibiotics doxycycline, chloramphenicol, tetracycline, trimethoprim–sulfamethoxazole, ciprofloxacin and nalidixic acid exceeded 85%.[17] In 2016, 144 isolates of V. cholerae O1 El Tor biotype, Ogawa serotype, from outbreaks in the provinces of Granma and Havana in June 2012 through January 2013 were studied. Isolates showed resistance values above 90% for trimethoprim–sulfamethoxazole, sulfonamide and ampicillin. Intermediate sensitivity was found to ciprofloxacin and chloramphenicol (30.6% and 27.1%, respectively). Sensitivity levels higher than 92% were observed for azithromycin, doxycycline, gentamicin and tetracycline. No multidrug-resistant strains were identified.[18]
Non-O1, non-O139 V. cholera is considered an international emerging pathogen. Although phenotypic characterization studies of these serogroups have been conducted since the 1980s, the relationship between virulence determinants and antimicrobial resistance in non-O1, non-O139 V. cholerae in Cuba is unknown. Because ADD caused by Vibrio species constitutes an important challenge for the Cuba’s health system, we set out to describe antimicrobial susceptibility to drugs commonly used in ADD treatment and verify the presence of enzymatic virulence factors.
METHODS
Design and population A cross-sectional descriptive study was conducted in January through November 2014, based on 125 non-O1, non-O139 V. cholerae isolates obtained from feces of patients with ADD in Cuba in 2013 and 2014. The isolates, kept in Pasteur conservation medium, belonged to the microbial culture collection of the National Reference Laboratory for Acute Diarrheal Diseases at IPK.
Variables Antimicrobial susceptibility was classified as sensitive, intermediate and resistant. Isolates were classified as sensitive if they were inhibited by antimicrobial concentrations reached at the recommended dose for the infection site. They were considered of intermediate sensitivity when they displayed minimum inhibitory concentrations close to the antimicrobial levels reached in blood or tissues with a lower degree of response than the sensitive strains. The intermediate category implies clinical efficacy in sites of the body where antimicrobials are physiologically concentrated or when the drug can be used in higher than normal doses. It also includes a buffer zone that could help avoid small technical factors (subjectivity in reading inhibition halos) that are difficult to control and may cause important discrepancies in interpretation, especially for drugs with narrow pharmacotoxic margins. Isolates were classified as resistant if they were not inhibited by serum antimicrobial concentrations achieved at standard doses or had minimal inhibitory concentrations in the range where specific resistance mechanisms are present and clinical efficacy is not demonstrable.
A resistance pattern was defined by resistance to one or more antimicrobial, of any family. MDR was defined by resistance to three or more antimicrobial families, as established by CLSI, 2017.[15] Virulence factors studied were beta-hemolysin, DNase, elastase, gelatinase and lecithinase.[11]
Antimicrobial families analyzed were beta-lactams (ampicillin), tetracyclines (doxycycline, tetracycline), sulfonamides (sulfonamide, trimethoprim–sulfamethoxazole), phenicols (chloramphenicol), macrolides (azithromycin), aminoglycosides (amikacin) and quinolones (ciprofloxacin).
Procedures Bacteriological identification was performed by conventional methods. Biochemical tests for the use of amino acids and carbohydrates for genus confirmation were done by the Moeller method. For species identification, isolates were tested for sodium chloride tolerance and sucrose uptake. Serological tests were performed by slide agglutination with O1 and O139 V. cholerae polyvalent antisera.[19]
The Bauer–Kirby agar diffusion method was used,[20] following CLSI 2010 criteria,[21] to test for isolates’ susceptibility to antimicrobials of choice and alternatives: ampicillin, doxycycline, tetracycline, sulfonamide, trimethoprim–sulfamethoxazole and chloramphenicol. Criteria established for the Enterobacteriaceae family were used for azithromycin and alternative antimicrobials such as amikacin and ciprofloxacin.[15]
Robinson’s technique was used to detect virulence factors beta-hemolysin, DNase, elastase and gelatinase,[22] Karagozova’s for enzyme lecithinase.[23]
Analysis Data were entered and processed using Excel. Absolute and relative frequencies were calculated for each of the study variables.
Ethics Data management procedures ensured confidentiality of patients whose isolates were tested. The study was approved by the IPK Ethics Committee.
RESULTS
Antimicrobial susceptibility Only 2 (1.6%) of the 125 isolates, showed sensitivity to all antimicrobials studied. Sensitivities of >96% were found to ciprofloxacin, doxycycline and azithromycin. Resistance was most frequent to ampicillin (60%), sulfonamides (46.4%) and trimethoprim–sulfamethoxazole (32%) (Table 1).
Resistance patterns Table 2 displays isolates’ distribution by resistance pattern. Six resistance patterns were found, four of them detected for the first time in Cuba (ampicillin + sulfonamide, ampicillin + tetracycline, ampicillin + ciprofloxacin, and sulfonamides + trimethoprim–sulfamethoxazole), as well as six MDR patterns (4.8%).
Virulence factors The proportions of virulence-factor positive isolates were: hemolysins 61.6% (77/125 isolates), DNase 60% (75), elastase 48.8% (61), gelatinase 90.4% (113) and lecithinase 77.6% (97).
Enzymatic virulence factors and resistance All monoresistant isolates expressed virulence factors, but proportions were smaller in isolates resistant to ≥2 microbials: hemolysins 66.3% (65/98), DNase 61.2% (60), elastase 46.9% (46), gelatinase 70.4% (69) and lecithinase 77.6% (76).
Table 1: Antimicrobial susceptibility of non-O1, non-O139 V. cholerae isolates (n = 125)
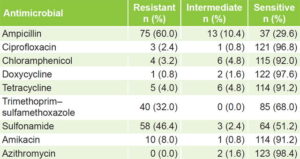
Table 2: Patterns of resistance and MDR of non-O1, non-O139 V. cholerae isolates (n = 125)
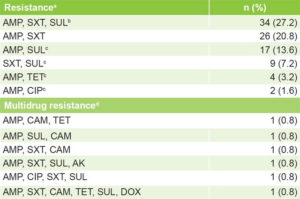
a resistance to ≥1 antimicrobials in any family b SXT and SUL belong to same family c new pattern dresistance to ≥3 antimicrobial families AMK: amikacin AMP: ampicillin CAM: chloramphenicol CIP: ciprofloxacin DOX: doxycycline MDR: multidrug resistance SUL: sulfonamide SXT: trimethoprim–sulfamethoxazole TET: tetracycline
DISCUSSION
Although this study found sensitivities exceeding 95% for the most widely used antibiotics for treatment of V. cholerae diarrhea (doxycycline, azithromycin and ciprofl oxacin), there is recent evidence of increased antimicrobial resistance among V. cholerae strains to commonly used drugs, implying serious treatment challenges worldwide.[24]
Our observed high rates of sensitivity to azithromycin echo previous findings reported in V. cholerae isolates from hospitals in Dhaka and Matlab, Bangladesh in 2000–2012, when 95% of isolates were azithromycin sensitive.[25] Mahmud’s study of isolates from a 2012 outbreak in Sierra Leone found 100% sensitivity to the same antimicrobial.[26] A publication based on testing 144 Cuban isolates of V. cholerae O1 from outbreaks reported 99.3% (143/144) sensitivity to azithromycin.[18] It is recommended as first-line therapy only for pediatric patients and pregnant women with suspected cholera, and as second-line in other cases.[27]
We found sensitivities to doxycycline similar to those described in Cuba by Fernández in 2016.[18] However, continued monitoring of its effectiveness in treating V. cholerae is important, since it is the first-line cholera treatment in Cuba and is also used for contact chemoprophylaxis.
Ciprofloxacin sensitivity was similar to that published by Mandal in India and Murhekar in New Guinea, 96.8% and 99%, respectively.[28,29] Sensitivities of 100% have been obtained in Cuba, by Bravo in 2006[8] and Cabrera in 2008.[17] These high sensitivities may be due to absence of mutations in the gyrA and parC genes or Cuba’s policy on treatment of ADD caused by V. cholerae, since ciprofloxacin is indicated in adult patients with compromised oral route or a history of allergy to the drug of choice (per MINSAP’s Cholera Control Program).[30]
The chloramphenicol sensitivities we observed were similar to those seen by Murhekar in New Guinea.[29] In Cuba, chloramphe-nicol is reserved for treatment of serious life-threatening infections, when there are no effective and less toxic treatment alternatives.[27] An interesting finding not previously reported in Cuba was identification of 4 (3.2%) isolates resistant to chloramphenicol. Thapa found 9.1% chloramphenicol resistance in Nepal in 2015.[6]
There was a high frequency of sensitivity to tetracycline, although somewhat less than the 100% published by Bakhshi in Iran in 2014.[31] In Cuba, Bravo found 98% of non-O1, non-O139 V. cholerae isolates were sensitive to tetracycline in a 10-year study (1996–2005).[8]
Aminoglycosides are a class of commonly used antimicrobials effective in clinical practice. Although there are several mechanisms, they are still active against the bulk of gram-negative aerobic bacilli, and amikacin can be used in cases of resistance.[27] The >90% sensitivity we found for amikacin is comparable to the 88.2% Rashed observed in Dhaka.[32] This is in sharp contrast to much lower sensitivity (8.3%) to amikacin in non-O, non-O139 V. cholerae reported by Bakhshi in Iran.[31]
Among the antimicrobials we studied, the highest percentage of resistance was observed for ampicillin. Bravo in Cuba and Dutta in India found 55.5% of V. cholerae isolates resistant to ampicillin,[9,33] slightly lower than the 60% observed in our study. In contrast, Thapa found 100% of O1 V. cholerae isolates resistant to this antimicrobial,[6] as did Shrestha in India and Talkington in V. cholerae isolates from Haiti.[34,35] These results could be due to production of extended-spectrum beta-lactamase; Gosh found this enzyme in 92.9% (52/56) of ampicillin-resistant V. cholerae isolates.[12] There is overwhelming evidence that emergence of antibiotic resistance is conditioned by selective pressure on the microbial flora because of inappropriate or excessive use of antibiotics.[36]
Four of the six resistance patterns we found were identified for the first time in Cuba. In 2006, Bravo’s Cuban study found some resistance patterns similar to those we observed (e.g., ampicillin + sulfonamide + trimethoprim–sulfamethoxazole),[8] and Bueno’s 2011 study (in Cuba) of 63 isolates of non-O1, non-O139 V. cholerae found that the predominant pattern was chloramphenicol + trimethoprim–sulfamethoxazole.[37] While the frequency of MDR isolates was low (<5%), in 2016, Fernández found no MDR.[18] Our finding of MDR could be due to the presence of integrons, natural gene acquisition systems that help bacteria capture exogenous genes and incorporate them into their genome, playing a prominent role in spread of resistance, because they often carry genes associated with mobile genetic elements.[12] Bakhshi suggests there may be class 1 integrons in enteropathogens.[31]
The presence of virulence factors may be due to their production by microorganisms of cellular proteins to invade the host immune system and achieve its colonization, since V. cholerae has several host colonization and infection mechanisms.[11] In addition to the cholera toxin, numerous extra and intracellular proteins are responsible for virulence, pathogenicity and cytotoxicity of V. cholerae O1, O139 and non-O1, non-O139.[17] In 2006, Cabrera (Cuba) found hemolysin and gelatinase in all isolates, DNase in 73.8%, lecithinase in 80%, and elastase in 86.1%.[17] Also in Cuba, Bueno found higher percentages than ours for gelatinase (96.8%) and hemolysin (92.1%), and lower for elastase (79.4%), lecithinase (73%) and DNase (68.3%).[37]
Each virulence factor was more frequent in monoresistant isolates than in those resistant to more than one antimicrobial. Other studies have related virulence to the presence of various virulence factors: Bina (USA) reported that V. cholerae isolates showing resistance–nodulation–division efflux pumps displayed decreased production of cholera toxin and pili coregulating toxin compared to wild-type strains, explained by reduced transcription of tcpP and toxT.[38] However, Spengler held that the presence of VexH efflux pumps in V. cholerae contributed to production of cholera toxin and pili coregulating toxin.[39]
It has been suggested that porin deficiency in Klebsiella pneumoniae increases antimicrobial resistance and decreases virulence. Similar results are reported for Neisseria meningitidis, Pseudomona aeruginosa, Vibrio spp and other Enterobacteriaceae. Studies suggest an inverse association between resistance and virulence, but the cause of this association is still unknown. Some authors suggest that it may be because less pathogenic serotypes maintain colonization for a longer time and are more exposed to multiple antimicrobials.[38,40,41]
One of the study’s limitations was lack of testing for minimal inhibitory concentration in resistant isolates. Nor was it possible to determine mechanisms of resistance in the isolates studied. Nevertheless, the results contribute to knowledge about the emergence and dissemination in Cuba of isolates resistant to various antimicrobials, and further our understanding of the determinants of virulence. It is advisable to maintain microbiological surveillance of non-O1, non-O139 V. cholerae in Cuba and to develop new studies to identify other virulence factors in these serogroups.
CONCLUSIONS
The results support continued use of the antimicrobials azithromycin, doxycycline and ciprofloxacin for treatment of infections caused by V. cholerae, and confirm the existence of four new resistance patterns in isolates circulating in Cuba.





