ABSTRACT
INTRODUCTION Approximately 73% of persons with HIV who receive antiretroviral therapy in Cuba are in viral suppression. The non-response of the remaining 27% could be due to several factors including adverse drug reactions and HIV resistance to antiretroviral drugs, as well as social factors and idiosyncratic characteristics of each patient. Genetic information explains from 20% to 95% of a drug’s effects and variations in response. Considering optimization of therapeutic efficacy in our country, genetic factors of the host should be identified.
OBJECTIVE Identify polymorphisms affecting genetic variability of responses to antiretroviral drugs.
EVIDENCE ACQUISITION A literature review was conducted (of original articles, published theses, clinical reports and bibliographic review studies, from 2000 to 2018, in Spanish and English listed in MEDLINE/PubMed, SciELO, LILACS, PharmGKB and Google Scholar) with the following key words: pharmacogenetics, human immunodeficiency virus, anti-retroviral agents, genetic polymorphism, genetic techniques, pharmacogenomic variants.
DEVELOPMENT The review identified 77 relevant publications meeting specific quality criteria. A summary table was built with data collected on antiretroviral drugs, genes and proteins involved in polymorphic variations, their associated effects and relevant scientific references. Information was included on polymorphisms related to 12 antiretroviral drugs used in HIV therapy. Polymorphisms determine variations in proteins involved in drug transport and metabolism and in elements of immunity. Relevant pharmacogenetic biomarkers recognized by drug regulatory agencies were identified.
CONCLUSIONS The study identified genetic variations (single-nucleotide polymorphisms) associated with 12 antiretroviral drugs. In most cases, no statistically significant causal association was found. Identifying polymorphic variations is a medium- and long-term objective that requires statistical support and adoption of strategies to optimize antiretroviral therapy. An approach combining plasma-level monitoring and pharmacogenetic analysis is recommended to optimize therapy for HIV patients.
KEYWORDS Pharmacogenetics, HIV, anti-retroviral agents, antiretroviral therapy, genetic polymorphism, genetic techniques, pharmacogenomic variants.
INTRODUCTION
HIV Infection has been a global health problem since 1983. At the end of 2016, approximately 36.7 million people worldwide were HIV-positive. In mid-2017, approximately 20.9 million people were receiving antiretroviral (ARV) drugs.[1] AIDS represents the infection’s advanced clinical stage. To treat HIV, numerous ARVs have been developed that act on different phases of viral replication and are used in combination as antiretroviral therapy (ART).
In Cuba, ARV drugs of all classes are used, although some of the latest-generation ARVs are only available in limited quantities and use of others has been discontinued due to their toxicity. Cuba’s ARV drugs are generic and produced domestically, procured by nongovernmental organizations through projects with The Global Fund to Fight AIDS, Tuberculosis and Malaria (Global Fund) or through personal donations. ART in Cuba meets national and international therapeutic standards. First-line treatment combines nucleoside reverse-transcriptase inhibitors (NRTIs) with non-nucleoside reverse transcriptase inhibitors (NNRTIs). Thanks to ARV use, incidence of HIV/AIDS-related deaths has fallen since 2005.[2] By December 31, 2018, in Cuba 31,118 persons had been diagnosed with HIV, 25,497 of whom were alive.[3]
IMPORTANCE: This study is an updated review of the pharmacogenetics of antiretroviral drugs. There have been few studies in Cuba on the subject of pharmacogenetics and no studies examining its effects on antiretroviral therapy. The study identified single nucleotide polymorphisms associated to 12 antiretroviral drugs. To optimize therapy, pharmacogenetic and plasma drug monitoring is recommended for HIV patients.
The Joint United Nations Programme on HIV/AIDS (UNAIDS) and its partners have set global goals for HIV diagnosis, treatment and viral suppression (known as 90–90–90): by 2020, at least 90% of people living with HIV should know their serological status, 90% of those diagnosed should be receiving sustained ART, and 90% of those receiving ART should have attained viral suppression (viral load <1000 copies/mL). Cuba shares these goals and in 2018 the country attained 83%–87%–73%,[3] meaning that 27% of patients receiving therapy did not attain viral suppression.
Factors responsible for non-response could include resistance to first-line ARV drugs (22% of patients), which has increased for NRTIs, NNRTIs, and protease inhibitors (PIs), to 52.7%, 54.7%, and 21.4%, respectively.[4] Adverse reactions (both moderate and severe) also can affect regimen adherence.[5] Alterations in blood bioavailability were reported in 25% of patients receiving the three classes of ARVs within their respective therapeutic windows.[6] The most common reason described in Cuban patients for changing an ART regimen was the appearance of an adverse drug reaction (ADR).[7]
Among the most serious ADRs reported in Cuba from 2003–2012 were two hepatotoxicity reactions associated with nevirapine and lamivudine, and a case of anemia due to zidovudine. No fatal reactions were reported during the period studied.[8] There is scant literature in Cuba on adverse reactions to ARV drugs, due partly to the low number of ADRs reported by health professionals.[8, 9] As the number of cases studied is small, ADR statistics may be underestimated. Cuba’s 2017 Annual Drug Safety Report,[2] lists 14 cases of ARV adverse reactions in a tertiary care institution, which represents 0.08% of all ADRs reported that year. The drugs cited in the drug safety reports were nevirapine (50% of reports), zidovudine (25%), abacavir (16.7%), and tenofovir (8.3%). Adverse drug reactions reported included headache, Stevens-Johnson syndrome, rash, vomiting, itchy skin and acute kidney failure.[2]
Current Cuban strategies to prevent therapeutic failure include surveillance of progress markers (viral load and T CD4+ lymphocytes) in clinical labs, and in some cases therapy monitoring, but the patient’s genetic factors are not addressed. Recent advances in pharmacogenetics (PGx) have made it possible to identify genetic variations that modulate response to these drugs and their toxicity. The main source of variability in human genomes are variations in a single nucleotide––adenine (A), thymine (T), cytosine (C), guanine (G)––known as single-nucleotide polymorphisms (SNPs).[10]
This study is an updated review of the PGx of ARV drugs. There have been few studies in Cuba on the subject of PGx and no studies examining its effects on ART. From the perspective of therapy optimization, we proposed to compile information on the main genetic polymorphisms described in the literature affecting response to ART and to identify useful strategies for their implementation in HIV therapy.
Evidence Acquisition Terms included in the information search: pharmacogenetics, HIV, anti-retroviral agents, antiretroviral therapy, genetic polymorphism, genetic techniques, pharmacogenomic variants. Bibliographic databases consulted: MEDLINE/PubMed, SciELO, LILACS and PharmGKB. Search engines: Google and Google Scholar. Types of documents: original articles, published theses, clinical reports and bibliographic reviews. Languages: Spanish and English. Dates of publication: 2000 to 2018. Exclusion criteria: no free access to complete text due to financial constraints, studies on genetic variations unrelated to HIV or presenting inadequate scientific evidence.
DEVELOPMENT
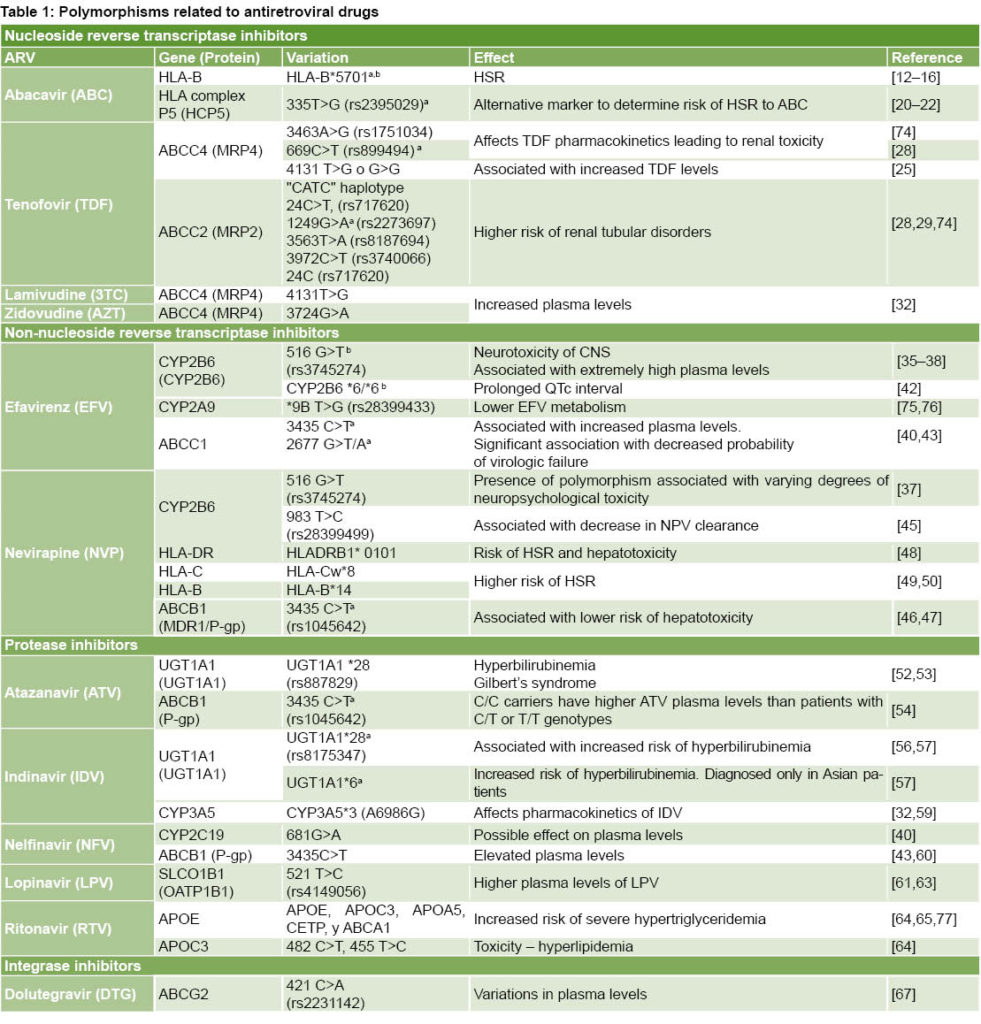
Of 1143 reports identified in the information search, 77 met the above-mentioned criteria. Table 1 summarizes information compiled on ARV drugs, genes and proteins associated to polymorphic variations and effects.
There are six classes of ARV drugs: nucleoside reverse-transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), entry inhibitors (EIs), fusion inhibitors (FIs), and integrase inhibitors (INIs). HIV therapy also uses post-attachment inhibitors (ibalizumab) and pharmacokinetic enhancers (cobicistat). Despite advances in patient survival and reduction of HIV-associated morbidity/mortality, adverse events and disorders associated with ARVs persist, which limit their benefits and contribute to drug resistance. The following is a discussion of updated information on the pharmacogenetics of ARV drugs based on literature review.
Nucleoside reverse-transcriptase inhibitors (NRTIs)
Abacavir (ABC): hypersensitivity reaction Since its introduction on the market, ABC has shown high efficacy and an acceptable toxicity profile for HIV treatment. About 5% of ABC-treated patients develop a hypersensitivity reaction (HSR) characterized by potentially fatal multisystemic effects. Symptoms generally appear within the first six weeks of treatment and range from fever, exanthema, and gastrointestinal symptoms to lethargy or general malaise. Symptoms generally worsen as treatment continues and improve within 24 hours of interruption.[11] One study reports this HSR occurring in 3.7% of ABC-treated patients and in up to 14% in some clinical trials.[12]
Although clinical symptoms are nonspecific and difficult to differentiate from other reactions, a close association has been described with presence of allele HLA-B*5701,[13,14] which can lead to activation of cytotoxic lymphocytes T CD8+, provoking secretion of two inflammation mediators––tumor necrosis factor (TNF)-alpha and interferon (IFN)-gamma––associated with appearance of delayed HSR.[13] The HLA-B*5701 allele is more prevalent in the Caucasian population (5% to 8%), less common among Japanese, Chinese and Koreans (< 1%), and rare in Sub-Saharan Africans. In South American Caucasians, prevalence is 5% to 7%.[14]
A 2002 study supporting the clinical value of identifying the HLA-B*57:01 allele found genotyping for HLA-B*5701 conducted in predominantly Caucasian populations had a positive predictive value of 100% for hypersensitivity to ABC, and absence of this allele combination had a negative predictive value of 97%.[13] In 2008, a prospective clinical trial revealed that identifying this allele (and the resulting modification of ARV therapy) was associated with a decline in incidence of hypersensitivity to ABC from 7.8% to 2.7%.[14] The HLA-B*5701 allele is identified through flow cytometry or sequencing with molecular amplification.[15] A skin test has been developed that has shown a satisfactory correlation between presence of HLA-B*5701 allele and risk of presenting HSR,[16] but as false negatives are not uncommon, the test’s use is not recommended in clinical practice.[17] This is among the earliest examples of PGx applied to clinical diagnosis in order to prevent drug-associated toxicity.
Some studies consider HLA-B*5701 genotyping cost-effective, while others argue that it is expensive, time consuming and requires a specialized laboratory, and thus propose another genetic marker, HCP5, instead.[18,19] Polymorphism 335T>G, located on gene P5 in the HLA complex HCP5, is in linkage disequilibrium with HLA-B*5701[20, 21] and its identification is easier, more economical and quicker. HCP5 SNP genotyping is being used more frequently as a simple method to detect possible hypersensitivity to ABC, although it presents some disadvantages.[22] In a study of 245 HIV patients, a close correlation was observed between HLA-B*5701 and HCP5 (negative and positive predictive values of 100% and 93%, respectively).[20]
Tenofovir disoproxil fumarate (TDF): renal toxicity TDF is generally well tolerated, with an excellent metabolic profile and no potential mitochondrial toxicity. Its most common ADR is renal toxicity, which is enhanced if co-administered with other drugs. The mechanism through which TDF causes renal tubular damage is not well understood; toxicity could be due to mitochondrial damage or interference with normal cell functions associated with intracellular drug accumulation.[23]
TDF is a substrate of transport proteins involved in its renal excretion, which occurs by both glomerular filtration and active tubular secretion. TDF enters proximal tubular cells by organic anion transporter (OAT) proteins. Its excretion in urine is mediated by multidrug-resistant protein MRP4, the main transporter of TDF at the apical level, which regulates TDF levels inside tubular cells.[24] Intervention of MRP7 and MRP2 proteins has also been described, although their role is unclear, as is the mechanism by which polymorphisms in MRP2 affect development of tubular dysfunction.[23] Polymorphisms in the genes encoding these proteins could alter their expression and activity, affecting TDF pharmacokinetics and favoring its accumulation in renal tubular cells. TDF-associated toxicity can produce proximal tubular dysfunction and acute tubular necrosis, which can progress to chronic kidney disease. Toxicity tends to reverse when the drug is withdrawn, but recovery may not be complete.[25] Nephrotoxicity in TDF patients is uncommon (about 2.5%),[26] but it must be taken into consideration given the growing use of this drug.
In gene ABCC4, which codes the MRP4 protein, three genetic polymorphisms were identified. Studies have shown that polymorphism 3463A>G is related to increased plasma and intracellular TDF levels as well as decreased renal clearance.[27] These results need to be confirmed, however, due to the small number of patients studied (n = 30). Polymorphism 669C>T in this gene was also identified; it is commonly found in patients who present renal tubular damage.[28] Another study concluded that after controlling for body weight, estimated glomerular filtration rate (GFR) and concomitant use of a PI reinforced with ritonavir, the average TDF plasma level in patients carrying genotype TG or GG ABCC4 4131 was about 30% higher than in patients who were carriers of TT.[25]
Polymorphisms in the ABCC2 gene that codes MRP2, specifically the CATC haplotype (combination of polymorphisms in positions -24C>T, 1249G>A, 3563T>A and 3972C>T) and -24C allele, have been associated with higher risk of tubulopathy.[28] According to Manosuthi, HIV patients who carry gene ABCC2 * genotype CC in position -24 or who present a high tenofovir plasma level have a higher risk of diminished GFR.[29] According to Izzedine,[28] genotype ABCC2-24CC and allele ABCC2-1249-A have been found more often in patients with tubular dysfunction than in patients with normal function. However, Rungtivasuwan[25] and Álvarez Barco[23] did not find that association. The most commonly observed SNPs in MPR2 play an indirect role: they affect transport of other substances that would influence TDF toxicity or by being in linkage disequilibrium with other SNPs which, when directly associated with expression of proteins involved in TDF transport, can lead to drug accumulation inside the cell, causing mitochondrial damage and/or interference with normal cell function. The most widely accepted hypothesis on the mitochondrial toxicity mechanism holds that it is due to inhibition of DNA polymerase γ, with a resulting decline in mitochondrial DNA required to maintain the electron transport chain proteins. This mitochondrial toxicity has been described for cidofovir and adefovir. Due to TDF’s structural similarity to these nucleotide analogues, it is speculated that TDF may also cause this type of cellular damage.[30]
Toxicity associated with TDF has been shown to be dependent on concentration, which would justify monitoring plasma TFV levels for pharmacokinetic studies of kidney function. However, there are drawbacks to doing so (intraindividual variability, lack of defined therapeutic range for TDF, etc.). Available information on genetic polymorphisms in renal toxicity risk with TDF therapies is contradictory and thus more evidence is needed. If prospective studies show these effects, patients at risk for developing tubular damage would be identified early. Use of pharmacogenetic markers together with quantification of TDF can alert physicians to the advisability of monitoring renal function.[23]
Lamivudine (3TC) and zidovudine (AZT): polymorphisms associated with increased plasma levels 3TC has high oral bioavailability, wide biodistribution and is generally well tolerated. Common adverse reactions include nausea, vomiting, diarrhea, stomach ache, hair loss, fever, insomnia, rash, nasal congestion and joint ache. AZT can cause serious adverse reactions such as hematological toxicity, including neutropenia and severe anemia, lactic acidosis and severe hepatomegalia with steatosis. Cases of hepatic decompensation have been reported for AZT and prolonged use has been associated with redistribution of body fat and lipodystrophy syndrome.[31]
A pilot study on the pharmacogenetic aspects of therapy with indinavir, AZT and 3TC in 33 adults living with HIV reported that the average AZT level was 49% higher in carriers of variant ABCC43724G˃A than in wild genotype (GG) carriers.[32] Carriers of variant 4131T>G of gene ABCC4 also showed a 20% increase in intracellular 3TC levels.
Few studies have evaluated pharmacogenetics with respect to AZT and 3TC, but the aforementioned results indicate that variations in gene coding for drug metabolism can influence ART efficacy and toxicity. Large-cohort studies are needed for more in-depth analysis and to validate the role of pharmacogenetic factors in AZT pharmacokinetics and pharmacodynamics.[33]
Non-nucleoside reverse transcriptase inhibitors (NNRTIs)
Efavirenz (EFV): polymorphisms in metabolizing enzymes and transport proteins that affect plasma levels EFV presents few clinically significant side effects, but frequently produces psychological and neurological symptoms, especially during the first three to four weeks of therapy. Not uncommonly, these symptoms cause discontinuation. This ARV drug has a narrow therapeutic window, since EFV plasma levels over 4 mg/mL have been associated with high toxicity in the central nervous system, and at levels below 1 mg/mL incidence of virologic failure appears to increase.[34]
EFV is metabolized by isoenzyme 2B6 of cytochrome P450 (CYP2B6) through primary oxidative hydroxylation.[34] Several polymorphisms have been associated with variations in this protein’s expression. The gene’s allelic variation that appears to have the biggest effect on hepatic expression of CYP2B6 and metabolism of EFV is the change of G to T in codon 516.[35-38] The effect of this change is decreased activity of the encoded protein; so carriers of this allele (especially homozygous persons) eliminate EFV more slowly, have higher EFV plasma levels[38] and higher incidence of especially intense and prolonged neuropsychological toxicity. The marked increase in EFV’s half-life increases the patient’s risk of developing resistance to the drug if it is suspended at the same time as all other ART components. In most of one study population, the polymorphism is associated with varying EFV plasma levels and central nervous system toxicities, suggesting that in patients presenting T/T genotype, prescribing lower EFV dosage could ensure reduced side effects without compromising drug efficacy,[39] but another study found no correlation between neurotoxicity and EFV plasma levels.[40] In any case, a combination of genotyping CYP2B6 and drug monitoring has been proposed to minimize toxicity and viral resistance.[41] Besides neurotoxicity, elevated EFV plasma levels have been associated with prolonged QTc interval in subjects with genotype CYP2B6 *6/*6, since they presented higher EFV levels in comparison with genotype *1/*1,[42] although the full potential of EFV to prolong the QTc interval may not be completely verified until conclusion of the study.[43]
Genetic polymorphisms of P glycoprotein (P-gp) also called multidrug resistance protein 1 (MDR1) have been widely studied. Among the most important are 3435 C>T and 2677 G>T/A, associated with decreased expression of the protein.[43] Several studies suggest that these polymorphisms could be related to low EFV levels. However, results are not conclusive.[41] One study notes that although these MDR1 variations have been significantly associated with resistance to EFV, exposure to these plasma levels is not the only pharmacological determinant of resistance, indicating the need for further studies to determine the mechanism by which an MDR1 polymorphism can affect virologic response.[40] According to Alonso-Villaverde[44] genotype C/C in codon 3435 of the MDR1 gene is associated with increased high-density lipoprotein (HDL) cholesterol levels in patients who receive EFV. This observation is supported by the fact that P-gp is involved in lipoprotein metabolism.
The fact that certain genetic polymorphisms may influence and condition significant differences in EFV pharmacokinetics between individuals could have important implications for effective ART. EFV is currently administered at a fixed dose of 600 mg daily. The possibility that a lower dose could reduce ADRs while sustaining efficacy in patients with allelic variations of CYP2B6 associated with higher exposure to the drug is attractive and has already been successfully applied in isolated cases.[41]
Nevirapine (NVP): hypersensitivity reaction NVP is metabolized mainly by enzymes CYP3A4 and CYP2B6, with a smaller contribution from CYP3A5. Several studies have reported that polymorphisms 516G>T[38] and 983T>C[45] in the CYP2B6 gene are associated with variations in NVP pharmacokinetics in different ethnic populations. Patients who are homozygous for this allelic variation presented NVP plasma levels 1.7 times higher than patients homozygous for the common allele.[37] Clinical implications of this observation are unclear, but since higher NVP plasma levels have been associated with greater risk of hepatic toxicity, patients with polymorphic allele 516G˃T may present elevated hepatic enzymes during NVP therapy.[39]
P-gp could be involved in transporting NVP outside cells. Reduced P-gp expression could cause NVP accumulation and higher plasma drug levels. Polymorphism 3435C>T in the MDR1 gene has been associated with low P-gp expression. In a study on patients in South Africa, this polymorphism was associated with lower risk of hepatotoxicity after starting NVP therapy.[46] Another report described the same observation, in which the T allele in MDR1 was associated with a lower risk of hepatotoxicity in patients receiving EFV or NVP.[47] A lower risk of hepatotoxicity in T/T carriers is paradoxical, since higher intracellular concentrations of NVP would be expected.[39]
Approximately 5% of patients treated with NVP experience an HSR consisting of rash and fever; hepatitis and other systemic symptoms also may occur occasionally, and in some cases can be fatal. Most side effects appear one to six weeks after start of therapy. The mechanism involved in appearance of NVP-related ADRs is not well understood. Skin reactions are most likely mediated by the major histocompatibility complex (MHC-I) and influenced by the CYP2B6 metabolism of NVP, while hepatic toxicity is most likely mediated by MHC-II and not affected by its metabolism. The simultaneous presence of allele HLA-DRB1*01:01 and more than 25% of LT-CD4+ significantly increases risk of HSRs and hepatotoxicity from NVP.[48]
Patients in Sardinia, Italy, who presented unusually high levels of HSRs to NVP compared with other ethnic groups were studied to see if there was a relationship with specific HLA antigens.[49] The study examined 49 HIV patients who were treated with NVP and 82 patients who were not. Approximately 26% of the patients receiving NVP (13/49) developed HSRs; of these, 46% (6/13) presented haplotypes HLA–Cw*8 and HLA–B*14, compared with only 5% (2/36) in the group who received NVP and did not develop HSR. Although the study was not able to determine which of the two haplotypes was more related to HSR, in a later study on a Japanese population where haplotype HLA–B*14 was not present, association between haplotype HLA–Cw*8 and risk of developing HSR due to NVP was confirmed.[50]
Protease inhibitors (PIs)
Atazanavir (ATV): hyperbilirubinemia ATV is included in first-line ART, both for patients initiating ART and those who have interrupted therapy due to intolerance of other ARVs. Atazanavir is metabolized primarily by isoenzyme CYP3A4 of cytochrome P450 and acts as an inhibitor of this isoenzyme and of others, such as uridine-glucuronosyltransferase 1, polypeptide A1 (UGT1A1).
One disadvantage associated with ATV use is that 20%–50% of patients treated with this drug develop hyperbilirubinemia, and about 6% present jaundice. Prevalence of the UGT1A1*28 variation (the most common polymorphism in the UGT1A1 superfamily) was found in 9%–60% of patients, with the highest prevalence in Africa.[51] This alteration does not have clinical consequences, but treatment is suspended in some patients, usually due to jaundice intensity. This ADR is attributed to competitive inhibition by ATV and the UGT1A1 enzyme responsible for conjugation and clearance of bilirubin and occurs more often (5%–10%) in the population with Gilbert syndrome (benign congenital disorder characterized by spontaneous increase in bilirubin plasma values). This syndrome is associated with the UGT1A1*28 allele, defined by the presence of seven repetitions of the dinucleotide TA (TA7) in the gene’s promoting region that codes the enzyme, which translates into reduced enzyme activity and asymptomatic hyperbilirubinemia.[52]
Another study confirmed the association between this allele and risk of hyperbilirubinemia; 67% of the individuals homozygous for the UGT1A1*28 allele who received ATV or IDV had at least two episodes of hyperbilirubinemia, compared with 7% in the group not treated with either of these drugs. The UGT1A1*28 allele is the cause of Gilbert syndrome. Prevalence of the UGT1A1*28 allele varies significantly between populations. Hyperbilirubinemia associated with atazanavir is more predictable if considered in conjunction with UGT1A1 genotypes, basal bilirubin and the initial hemoglobin values.[53]
A study in Spain on 118 patients treated with ATV enhanced with ritonavir (RTV) showed a correlation between bilirubin values and ATV plasma levels, and a relationship between ATV plasma levels and polymorphism 3435C>T in the MDR1 gene that codes P-gp.[54] This is the same profile observed in Caucasian patients with genotype CT/TT, who have lower ATV levels even when ATV is enhanced with RTV. Although ATV plasma levels are directly related to bilirubin levels, risk of severe hyperbilirubinemia increases even more in the presence of the UGT1A1*28 (TA7) allele. In the study, C/C carriers had higher ATV plasma levels than patients with C/T or T/T genotype, which suggests that P-gp plays an important role in the bioavailability of ATV. The change in nucleotide C for T in position 3435 and in the MDR1 gene is silent (does not generate phenotypic changes) so it does not change the coded amino acid.[55] Consequently, this may be a marker for another P-gp gene polymorphism with functional effects.
Differences in ATV plasma levels in patients with different P-gp genotypes could be particularly relevant and some patients may need higher ATV plasma levels to inhibit replication of the virus. In such cases, patients with genotype MDR1 CC could have a pharmacokinetic advantage over patients with other genotypes. These studies also show the clinical value of the UGT1A1 test before prescribing ATV, at least in Caucasian patients.[54]
Indinavir (IDV): hyperbilirubinemia IDV is another PI that is not currently widely used due to its difficult dose prescription and its narrow therapeutic index. From 5% to 25% of patients who receive it develop hyperbilirubinemia through the same mechanism as atazanavir.[17] Patients with the additional dinucleotide (TA) present in allele *28 have increased bilirubin levels compared with those who do not have the polymorphic allele.[56] A study in Thailand of patients treated with IDV suggests that a different allele, UGT1A1*6, can predispose a patient to hyperbilirubinemia more than the UGT1A1*28 allele.[57]
IDV is metabolized mainly in the liver by isoenzyme CYP3A5, which shows wide genetic variation. The *3 allele is one of the best known in CYP3A5; its prevalence varies with ethnicity .[58] A guanine in position 6986 in this allele creates a cryptic splicing site in intron 3, which leads to an aberrant splicing in the majority of transcriptions and reduced expression of CYP3A5 protein.[59] Presence of this variation has been associated with low or null expression of CYP3A5 protein, and thus, low IDV clearance. As a result, individuals with this polymorphism tend to present higher IDV plasma levels and more commonly suffer from kidney stones (nephrolithiasis) in IDV-containing therapy. Although these data can appear clinically insignificant due to the benign nature of increased bilirubin, such findings illustrate an important point: genetic variability in pharmacological targets affects drug response.[32]
Nelfinavir (NFV): polymorphisms that affect its plasma levels Metabolism of NFV is mainly hepatic due to the CYP450 pathway. Enzymes CYP3A and CYP2C19 appear to be predominant in humans. An association has been noted between 681G>A (CYP2 C19*2) and NFV plasma concentrations.[40] Individuals who are heterozygous or homozygous for the rare allele present higher concentrations of this PI than individuals homozygous for the common allele. The same study also showed that heterozygous individuals have lower NFV levels.
NFV plasma levels are influenced by polymorphisms in the MDR1 gene. A correlation between NFV plasma levels and genotypes in position 3435 has been described.[43] A study described lower levels in patients carrying the T/T variation, although carriers of the T/T allele have reportedly shown higher levels of T CD4 cells when receiving NFV therapy, despite its unfavorable pharmacokinetic profile. However, this observation has not been confirmed by other authors.[39] Another study showed that heterozygous (C/T) children who are carriers of polymorphism 3435 had higher NFV plasma levels than children with C/C or T/T genotypes.[60] The researchers measured intracellular NFV levels in lymphoblastoid cell lines and examined genotypes of the 3435 position, finding significantly higher NFV levels in the T/T genotype than in the C/C or C/T genotypes. These data support the prior observation of better immunologic recovery in NFV therapy for T/T carriers. It is not clear whether presence of the T allele is associated with higher or lower NFV plasma levels. Differences between the two studies [39,60] e.g., age of population studied and concomitant drugs that may influence P-gp activity, could explain differing results.
Lopinavir (LPV): polymorphisms associated with increased plasma levels One study showed significant association of polymorphism SLCO1B1 T521C with higher LPV plasma levels in patients who are homozygous for the mutant allele,[61] which would suggest that LPV’s entry into the liver via transporter SLCO1A2 is an important determinant of exposure to LPV. However, no significant associations were observed between LPV levels and polymorphisms of genes SLCO1A2 and SLO1B3.[62]
Another study examined the influence of several genetic variations of gene SLCO1B, showing higher LPV plasma levels in carriers of mutant allele (521C) than in patients who were homozygous for the wild type allele (521TT). Reduced LPV capture by hepatocytes in carriers of allele 521C could explain these results. However, additional studies are needed to confirm clinical importance of SLCO1B1 polymorphisms in LPV pharmacokinetics.[63]
Ritonavir (RTV): dyslipidemia RTV is widely used at low doses as an enhancer of other PIs, increasing their plasma levels due to its inhibitor effect on the cytochrome P450 enzymatic system. Serious adverse reactions to RTV include possible metabolic toxicity and its effect on adipose tissue due to possible long-term consequences for cardiovascular risk.
A longitudinal cohort study of 329 patients followed for 3 years in Switzerland confirmed an association between SNPs of C-III (APOC3) apolipoprotein and hyperlipidemia. A group of patients carrying both variations of APOC3 and APOE (5.8%) presented higher risk of severe hypertriglyceridemia from treatment with RTV.[64]
A 4-year longitudinal study of 438 HIV-1 patients examining the effect of 20 SNPs on 13 genes associated with dyslipidemia in the general population suggests participation of other genes in lipidic ART response.[65] The objective was to help predict which patients were at risk for developing secondary dyslipidemia from ART. The study concluded that variations in 5 genes (ABCA1, APOA5, APOC3, APOE and CETP) helped explain triglyceride and cholesterol plasma levels, especially in the ART context. However, these findings lack current clinical applicability because of the partially unknown polygenic nature of the mechanism involved.
Variations in the APOC3 allele and its genes appear to contribute significantly to increased triglyceride plasma levels, more or less equal in measure to factors such as age and RTV use. Polymorphisms in gene APOC3 associated with higher risk of hyperlipidemia include -482C˃T, -455T˃C and 3238C˃G.[64] Foulkeset al. found that race/ethnicity was a predictor of plasma lipids in HIV-1 patients on ART and a significant difference in the influence of apoC-III genotypes on PI use across ethnic groups in the association with triglycerides.[66]
Integrase inhibitors (INIs)
Dolutegravir (DTG): polymorphisms that can affect DTG plasma levels Dolutegravir is a potent and well-tolerated INI drug. It is metabolized in the liver via the uridine diphosphate glucuronosyltransferase 1 (UGT1A1) family and cytochrome P450 (CYP3A4) family.
Genetic polymorphisms of ABCG2 were associated with maximum DTG plasma levels.[67] The genetic polymorphism of ABCG2 altered the protein’s expression level in experiments on plasmid transfection. Substitution of nucleotide C for A in position 421 significantly reduced the protein’s expression and activity. This genotype’s low expression level can help explain the high DTG levels, because absorption of DTG from intestinal lumen to capillaries increases in the presence of low expression of ABCG2 in the intestines. The authors propose that it is possible to predict high DTG plasma levels by identifying genotype before starting DTG‑containing therapy. In ABCG2 patients, DTG dose can be lowered to minimize possible toxicity and cost without compromising its potency.[67]
Summary Analysis
In summary, a literature review identified 12 ARV drugs with therapeutic responses affected by genetic polymorphisms. To facilitate application by health professionals working with HIV patients, Table 1 summarizes data obtained on these 12 drugs and the associated polymorphisms and effects.
In the drug safety studies consulted,[5,8,68] an HSR to abacavir (ABC) was not among the most frequently reported ADRs in Cuba; this could be due as much to underreporting of ADRs to ARV drugs as to the infrequent prescription of ABC.[5] In treatment-naive patients, therapy preferably begins with 3TC, AZT and NVP. In the Pedro Kourí Tropical Medicine Institute, various cases of rashes and Stevens Johnson syndrome associated with NVP use have been reported.[68] The authors suggest that this could be because HIV patients have higher exposure to medications and pronounced immunosuppression, as well as coinfection with other viral agents that could stimulate the immune system and predispose them to ADRs. However, appearance of these ADRs could also result in patients carrying the alleles associated with them––an association yet to be demonstrated.
The presence of genetic polymorphisms in metabolizing enzymes or transport/receptor proteins in certain individuals affects the PK/PD processes of some ARV drugs.[39,62,69] Increased ARV plasma levels can have various side effects, including rash,[13] anemia,[32,33] and hepatotoxicity.[38,70] When concentrations drop to suboptimal levels, viral suppression is incomplete and selective pressure triggers viral resistance to that drug. For this reason, we recommend monitoring blood levels in Cuban HIV patients who present ADRs or viral resistance to determine if ARV levels are within the therapeutic window and to adjust the dosage accordingly.
Most ARV drugs belong to the PI family. This group of drugs is one of the most widely used clinically. PIs are generally enhanced with RTV due partly to that drug’s high genetic barrier for developing resistance (meaning that accumulation of multiple mutations is needed to induce resistance). Patients with HIV receiving PIs as part of their ART are at high risk of suffering dyslipidemias and lipodystrophy syndrome. This association is relevant because these ADRs are among those often associated with treatment interruption and they occur frequently in Cuba.[68]
In their drug labelling, the US Food and Drug Administration (FDA), European Medicines Agency (EMA), and Japan’s Pharmaceuticals and Medical Devices Agency (PMDA) now include information concerning the following pharmacogenetic biomarkers:
—HLA-B for ABC, due to its proven association with appearance of an HSR. It is one of the most cited examples in the literature consulted. Before starting treatment with ABC, all patients should be tested to detect the HLA-B * 5701 allele as this drug is contraindicated in positive cases. Even if the association is negative or cannot be determined, suspension of the drug is clearly indicated after any hypersensitivity reaction to ABC appears, since its continuation could lead to more severe symptoms or even death;[71]
—CYP2B6 for EFV due to association in carriers of CYP2B6 *6/*6 genotype with high EFV plasma levels and prolonged QTc interval, according to the FDA;
—CYP2B6 516 G˃T (rs3745274) genotype for EFV due to recognized association with increased EFV plasma levels, according to the PMDA and EMA; and
—UGT1A1 for DTG since carriers of this polymorphism tend to have higher plasma DTG levels, according to the FDA. In the literature consulted, however, Chen[72] concluded that the association is not clinically significant.
ARV: Antiretroviral ART: Antiretroviral therapy ADR: Adverse drug reaction PK: Pharmacokinetics PD: Pharmacodynamics
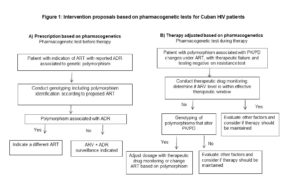
In current clinical practice abroad, specifically in management of HIV patients, most drugs are submitted to dosage adjustments a priori based on each patient’s genetic information and a posteriori based on the patient’s response. However, in light of therapeutic failure rates, new strategies are needed to optimize ART. Figure 1 shows two proposed interventions combining plasma level monitoring and pharmacogenetic analysis. The purpose of the first intervention (A) is to determine the most appropriate prescription according to a patient’s genetic information and is a useful guide for preventing the severe ADRs reported in therapeutic guidelines and drug safety studies. Based on the scientific evidence, we recommend that this strategy be implemented in patients prescribed ART containing NVP or ABC. The second recommended action (B)––applicable to patients receiving ART who present therapeutic failure and whose resistance test was negative––is to conduct therapeutic drug monitoring and identify polymorphisms to determine if a dosage should be adjusted or therapy changed. The triggering ARV drugs in this case are EFV and TDF, due to the numerous dosing regimens proposed, associated toxicities and potential interactions, together with the patient’s noncompliance with the therapeutic regimen. Incorporating these recommendations into treatment implementation protocols requires a laboratory with molecular biology capacity, drug quantification and specialized personnel to conduct the proposed diagnostics.
In Cuba, these biomarkers could be introduced in clinical practice according with the government’s newest set of guidelines for pharmacogenomics studies enacted in 2018.[73] The first recommendation is to study the incidence of adverse events and therapeutic failures associated with ARV drugs, since current statistics are inaccurate due to underreporting. Also, findings from this review have led to recommendations for clinical practice innovation to combine plasma level monitoring and pharmacogenetic analysis. We recommend that ARV plasma levels be monitored to help estimate the extent of influence of polymorphisms in ARV efficacy and that operational and cost-benefit studies be conducted to measure the impact and feasibility of this type of diagnosis. Incorporating these measures into clinical practice could help extend the therapeutic index of ARV treatment regimens, minimize the likelihood of developing drug resistance and ADRs, and result in a sustained response that improves patient quality of life and helps attain viral suppression in 90% of patients treated, thus reducing HIV transmission.
CONCLUSIONS
Twelve ARV drugs were identified that have been studied for their possible association with genetic variations (SNPs) in several genes, although in most cases there was no statistical support for a causal association.
To the extent that statistical analysis supports associations between these SNPs, clinical response and other pharmacological factors, combining drug monitoring with the identification of polymorphic variations before or during therapy is a medium- to long-term strategy to optimize ART.
References

- WHO. World health statistics: Monitoring health for the SDGs [Internet]. Geneva: World Health Organization; 2018 Jun 6 [cited 2018 Jul 6]. 86 p. Available from: https://www.who.int/gho/publications/world_health_statistics/2018/en/
- Ministry of Public Health (CU). Reporte Annual de Farmacovigilancia 2017 [Internet]. Havana: Ministry of Public Health (CU); 2017 [cited 2018 Apr 6]. 5 p. Available from: https://www.cecmed.cu/sites/default/files/adjuntos/vigilancia/farmacov/reporte_farmacovigilancia_2017.pdf. Spanish.
- Ministry of Public Health (CU). Informe anual VIH/Sida 2018. Havana: Ministry of Public Helath (CU); 2018. Spanish.
- Alemán Campos Y, Kouri V, Pérez Santos L, Fonseca C, Pérez JM, Ortega L, et al. HIV-1 Antiretroviral Resistance in Cuba, 2009–2014 [Internet]. MEDICC Rev 2018 Jul [cited 2019 Apr 6];20(3):15–21. Available from: https://scielosp.org/pdf/medicc/2018v20n3/15-21/en. Spanish.
- Alfonso Orta I, Toledo Romaní ME, Coutín Marie G, García Fariñas A, González Cruz R, López Jiménez G. Reacciones adversas a los antimicrobianos en Cuba (2003-2012). Rev Cubana Salud Pública [Internet]. 2016 Apr–Jun [cited 2019 Jun 20];42(2):294–305. Available from: http://scielo.sld.cu/scielo.php?script=sci_arttext&pid=S0864-34662016000200012. Spanish.
- Duménigo A. Cuantificación de antirretrovirales en sangre y análisis de efectividad en pacientes cubanos con VIH/sida tratados con zidovudina/lamivudina/nevirapina [thesis]. [Havana]: University of Havana, Institute of Pharmacology and Food; 2017. Spanish.
- Hernández Requejo D, Pérez Ávila J. Causas de cambio de tratamiento en un grupo de pacientes VIH/SIDA cubanos. Rev Cubana Invest Bioméd [Internet]. 2010 Apr–Jun [cited 2019 Mar 13];29(2):223–30. Available from: http://scielo.sld.cu/scielo.php?script=sci_arttext&pid=S0864-03002010000200007&nrm=iso. Spanish.
- Alfonso Orta I. Reacciones adversas a los antimicrobianos. Sistema Cubano de Farmacovigilancia, 2003–2012 [thesis]. [Havana]: Dr Pedro Kourí Institute of Tropical Medicine; 2014. Spanish.
- Frómeta Báez M, Alfonso Orta I, Martínez Rodríguez A. Reacciones adversas a los antirretrovirales en pacientes VIH/SIDA. Instituto Pedro Kourí, 2015. Rev Cubana Tecnol Salud [Internet]. 2017 [cited 2019 Mar 13];8(1):12–8. Available from: http://www.medigraphic.com/pdfs/revcubtecsal/cts-2017/cts171c.pdf. Spanish.
- International HapMap Consortium. The International HapMap Project. Nature [Internet]. 2003 Dec 18 [cited 2019 Mar 13];426(6968):789–96. Available from: https://doi.org/10.1038/nature02168
- Hetherington S, McGuirk S, Powell G, Cutrell A, Naderer O, Spreen B, et al. Hypersensitivity reactions during therapy with the nucleoside reverse transcriptase inhibitor abacavir. Clin Ther [Internet]. 2001 Oct [cited 2018 Jan 5];23(10):1603–14. Available from: https://www.ncbi.nlm.nih.gov/pubmed/11726000
- Hewitt RG. Abacavir hypersensitivity reaction. Clin Infect Dis [Internet]. 2002 Apr 15 [cited 2019 Mar 10];34(8):1137–42. Available from: https://academic.oup.com/cid/article/34/8/1137/284013
- Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002 Mar 2;359(9308):727–32.
- Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008 Feb 7;358(6):568–79.
- Gatanaga H, Honda H, Oka S. Pharmacogenetic information derived from analysis of HLA alleles. Pharmacogenomics. 2008 Feb;9(2):207–14.
- Saag M, Balu R, Phillips E, Brachman P, Martorell C, Burman W. High sensitivity of human leukocyte antigen-b*5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin Infect Dis [Internet]. 2008 Apr 1 [cited 2019 Mar 10];46(7):1111–8. Available from: https://www.ncbi.nlm.nih.gov/pubmed/18444831
- U.S. Department of Human Resources [Internet]. Washington, D.C.: U.S. Department of Human Resources; c2019. HIV/AIDS News. Updated Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents; 2009 Dec 1 [cited 2019 Mar 13]; [about 2 screens]. Available from: https://aidsinfo.nih.gov/contentfiles/lvguidelines/AdultandAdolescentGL.pdf
- Hughes DA, Vilar FJ, Ward CC, Alfirevic A, Park BK, Pirmohamed M. Cost-effectiveness analysis of HLA B*5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics [Internet]. 2004 Jun [cited 2019 Jan 5];14(6):335–42. Available from: https://www.ncbi.nlm.nih.gov/pubmed/15247625
- Nieves Calatrava D, Calle-Martín Ode L, Iribarren-Loyarte JA, Rivero-Román A, García-Bujalance L, Pérez-Escolano I, et al. Cost-effectiveness analysis of HLA-B*5701 typing in the prevention of hypersensitivity to abacavir in HIV+ patients in Spain. Enferm Infecc Microbiol Clin [Internet]. 2010 Nov [cited 2019 Jan 7];28(9):590–5. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0213-005X(10)00008-X
- Rodríguez-Nóvoa S, Cuenca L, Morello J, Córdoba M, Blanco F, Jiménez-Nácher I, et al. Use of the HCP5 single nucleotide polymorphism to predict hypersensitivity reactions to abacavir: correlation with HLA-B*5701. J Antimicrob Chemother [Internet]. 2010 Aug [cited 2019 Mar 13];65(8):1567–9. Available from: https://academic.oup.com/jac/article-lookup/doi/10.1093/jac/dkq204
- Sánchez-Girón F, Villegas-Torres B, Jaramillo-Villafuerte K, Silva-Zolezzi I, Fernández-López JC, Jiménez-Sánchez G, et al. Association of the genetic marker for abacavir hypersensitivity HLA-B*5701 with HCP5 rs2395029 in Mexican Mestizos. Pharmacogenomics [Internet]. 2011 Jun [cited 2019 Mar 13];12(6):809–14. Available from: http://www.futuremedicine.com/doi/full/10.2217/pgs.11.31?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed
- Colombo S, Rauch A, Rotger M, Fellay J, Martínez R, Fux C, et al. The HCP5 single-nucleotide polymorphism: a simple screening tool for prediction of hypersensitivity reaction to abacavir. J Infect Dis [Internet]. 2008 Sep 15 [cited 2018 Dec 12];198(6):864–7. Available from: https://academic.oup.com/jid/article/198/6/864/914628
- Álvarez Barco E. Estudio de factores farmacogenéticos y farmacocinéticos asociados con alteraciones renales en pacientes VIH positivos a tratamiento con tenofovir [thesis]: [Madrid]: Universidad Complutense de Madrid; 2014 [cited 2019 Mar 13]. Available from: https://dialnet.unirioja.es/servlet/tesis?codigo=100452. Spanish.
- Uwai Y, Ida H, Tsuji Y, Katsura T, Inui K. Renal transport of adefovir, cidofovir, and tenofovir by SLC22A family members (hOAT1, hOAT3, and hOCT2). Pharm Res [Internet]. 2007 Apr [cited 2019 Mar 10];24(4):811–5. Available from: https://doi.org/10.1007/s11095-006-9196-x2
- Rungtivasuwan K, Avihingsanon A, Thammajaruk N, Mitruk S, Burger DM, Ruxrungtham K, et al. Influence of ABCC2 and ABCC4 polymorphisms on tenofovir plasma concentrations in Thai HIV-infected patients. Antimicrob Agents Chemother [Internet]. 2015 [cited 2018 Mar 15];59(6):3240–5. Available from: http://aac.asm.org/cgi/pmidlookup?view=long&pmid=25801567
- Madeddu G, Bonfanti P, De Socio GV, Carradori S, Grosso C, Marconi P, et al. Tenofovir renal safety in HIV-infected patients: results from the SCOLTA Project. Biomed Pharmacother [Internet]. 2008 Jan [cited 2018 Mar 10];62(1):6–11. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0753-3322(07)00088-1
- Kiser JJ, Aquilante CL, Anderson PL, King TM, Carten ML, Fletcher CV. Clinical and genetic determinants of intracellular tenofovir diphosphate concentrations in HIV-infected patients. J Acquir Immune Defic Syndr [Internet]. 2008 Mar 1[cited 2018 Mar 10];47(3):298–303. Available from: http://insights.ovid.com/pubmed?pmid=18398970
- Izzedine H, Hulot JS, Villard E, Goyenvalle C, Domínguez S, Ghosn J, et al. Association be tween ABCC2 gene haplotypes and tenofovir-induced proximal tubulopathy. J Infect Dis [Internet]. 2006 Dec 1 [cited 2019 Apr 12];194(11):1481–91. Available from: https://academic.oup.com/jid/article-ookup/doi/10.1086/508546
- Manosuthi W, Sukasem C, Thongyen S, Nilkamhang S, Sungkanuparph S. ABCC2*1C and plasma tenofovir concentration are correlated to decreased glomerular filtration rate in patients receiving a tenofovir-containing antiretroviral regimen. J Antimicrob Chemother [Internet]. 2014 Aug [cited 2018 Dec 5];69(8):2195–201. Available from: https://academic.oup.com/jac/article-lookup/doi/10.1093/jac/dku129
- ADHOC International Steering Committee. A randomized placebo-controlled trial of adefovir dipivoxil in advanced HIV infection: the ADHOC trial. HIV Med [Internet]. 2002 Oct [cited 2018 Dec 5];3(4):229–38. Available from: https://onlinelibrary.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=1464-262&date=2002&volume=3&issue=4&spage=229
- Vadecum [Internet]. Madrid: Vadecum; c2019. Pediamécum. Edición 2015. Zidovudina; 2015 [cited 2019 Mar 6]. Available from: https://www.vademecum.es/medicamento-flexible-dermatix-26622
- Anderson PL, Lamba J, Aquilante CL, Schuetz E, Fletcher CV. Pharmacogenetic characteristics of Indinavir, Zidovudine, and Lamivudine therapy in HIV-infected adults: a pilot study. J Acquir Immune Defic Syndr [Internet]. 2006 Aug 1 [cited 2019 Mar 13];42(4):441–9. Available from: http://insights.ovid.com/pubmed?pmid=16791115
- Ghodke Y, Anderson PL, Sangkuhld K, Lamba J, Altman RB, Klein TE. PharmGKB summary: zidovudine pathway. Pharmacogenet Genomics [Internet]. 2012 Dec [cited 2019 Mar 13];22(12):891–4. Available from: http://insights.ovid.com/pubmed?pmid=22960662
- Marzolini C, Telenti A, Decosterd LA, Greub G, Biollaz J, Buclin T. Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV-1-infected patients. AIDS [Internet]. 2001 Jan 5 [cited 2019 Mar 13];15(1):71–5. Available from: http://insights.ovid.com/pubmed?pmid=11192870
- Gatanaga H, Hayashida T, Tsuchiya K, Yoshino M, Kuwahara T, Tsukada H. Successful efavirenz dose reduction in HIV type 1-infected individuals with cytochrome P450 2B6 *6 and *26. Clin Infect Dis [Internet]. 2007 Nov [cited 2019 Mar 13];45(9):1230–7. Available from: https://academic.oup.com/cid/article-lookup/doi/10.1086/522175
- Nolan D, Phillips E, Mallal S. Efavirenz and CYP2B6 polymorphism: implications for drug toxicity and resistance. Clin Infect Dis [Internet]. 2006 Feb 1 [cited 2019 Mar 13];42(3):408–10. Available from: https://academic.oup.com/cid/article-lookup/doi/10.1086/499369
- Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics [Internet]. 2005 Jan [cited 2019 Mar 13];15(1):1–5. Available from: http://insights.ovid.com/pubmed?pmid=15864119
- Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics [Internet]. 2007 Jun [cited 2018 Mar 20];8(6):547–58. Available from: http://www.futuremedicine.com/doi/full/10.2217/14622416.8.6.547?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed
- Rodríguez-Nóvoa S, Barreiro P, Jiménez-Nácher I, Soriano V. Overview of the pharmacogenetics of HIV therapy. Pharmacogenomics J [Internet]. 2006 Jul–Aug [cited 2019 Mar 13];6(4):234–45. Available from: http://dx.doi.org/10.1038/sj.tpj.6500374
- Haas DW, Smeaton LM, Shafer RW, Robbins GK, Morse GD, Labbe L, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing Efavirenz and/or Nelfinavir: an Adult Aids Clinical Trials Group Study. J Infect Dis [Internet]. 2005 [cited 2019 Jul 8];192(11):1931–42. Available from: https://academic.oup.com/jid/article-lookup/doi/10.1086/497610
- Sánchez Martin A. Terapia personalizada en la infección por el VIH: aplicación de criterios farmacocinéticos y farmacogenéticos [thesis]. [Salamanca]: Universidad De Salamanca, Facultad De Farmacia Departamento de Farmacia y Tecnología Farmacéutica; 2010 [cited 2019 Mar 13]. Available from: https://books.google.com.cu/books/about/Terapia_personalizada_en_la_infecci%C3%B3n_p.html?id=PqF7Uxcnc60C&redir_esc=y. Spanish.
- Abdelhady AM, Shugg T, Thong N, Lu JB, Kreutz Y, Jaynes HA, et al. Efavirenz Inhibits the Human Ether-A-Go-Go Related Current (hERG) and Induces QT Interval Prolongation in CYP2B6*6*6 Allele Carriers. J Cardiovasc Electrophysiol [Internet]. 2016 Oct;27(10):1206–13. Available from: https://doi.org/10.1111/jce.13032
- Fellay J, Marzolini C, Meaden ER, Back DJ, Buclin T, Chave JP, et al. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet [Internet]. 2002 Jan 5 [cited 2019 Mar 13];359(9300):30–6. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0140-6736(02)07276-8
- Alonso-Villaverde C, Coll B, Gómez F, Parra S, Camps J, Joven J, et al. The efavirenz-induced increase in HDL-cholesterol is influenced by the multidrug resistance gene 1 C3435T polymorphism. AIDS [Internet]. 2005 Feb 18 [cited];19(3):341–2. Available from: http://insights.ovid.com/pubmed?pmid=15718846
- Dickinson L, Chaponda M, Carr DF, van Oosterhout JJ, Kumwenda J, Lallo DG, et al. Population pharmacokinetic and pharmacogenetic analysis of nevirapine in hypersensitive and tolerant HIV-infected patients from Malawi. Antimicrob Agents Chemother [Internet]. 2014 [cited 2019 Mar 10];58(2):706–12. Available from: http://aac.asm.org/cgi/pmidlookup?view=long&pmid=24217698
- Haas DW, Bartlett JA, Andersen JW, Sanne I, Wilkinson GR, Hinkle J, et al. Pharmacogenetics of nevirapine-associated hepatotoxicity: an Adult AIDS Clinical Trials Group collaboration. Clin Infect Dis [Internet]. 2006 Sep 15 [cited 2019 Mar 10];43(6):783–6. Available from: https://academic.oup.com/cid/article-lookup/doi/10.1086/507097
- Ritchie M, Motsinger A, Donahue J, Erdem H, Raffanti S. Genetic variation in drug transporter and metabolizing enzyme genes may be associated with non-nucleoside reverse transcriptase inhibitor hepatotoxicity. Presentation at the12th Conference on Retroviruses and Opportunistic Infections; Boston; 2005.
- Phillips E, Bartlett JA, Sanne I, Lederman MM, Hinkle J, Rousseau F, et al. Associations between HLA-DRB1*0102, HLA-B*5801, and hepatotoxicity during initiation of nevirapine-containing regimens in South Africa. J Acquir Immune Defic Syndr [Internet]. 2013 Feb 1 [cited 2018 Jul 6];62(2):55–7. Available from: http://insights.ovid.com/pubmed?pmid=23328091
- Littera R, Carcassi C, Masala A, Piano P, Serra P, Ortu F, et al. HLA-dependent hypersensitivity to nevirapine in Sardinian HIV patients. AIDS [Internet]. 2006 Aug 1 [cited 2018 Jul 6];20(12):1621–6. Available from: http://insights.ovid.com/pubmed?pmid=16868443
- Gatanaga H, Yazaki H, Tanuma J, Honda M, Genka I, Teruya K, et al. HLA-Cw8 primarily associated with hypersensitivity to nevirapine. AIDS [Internet]. 2007 Jan 11;21(2):264–5. Available from: https://journals.lww.com/aidsonline/Fulltext/2007/01110/HLA_Cw8_primarily_associated_with_hypersensitivity.29.aspx
- Panagopoulos P, Maltezos E, Hatzakis A, Paraskevis D. Hyperbilirubinemia in atazanavir treated HIV-infected patients: the impact of the UGT1A1*28 allele. Pharmgenomics Pers Med [Internet]. 2017 Jun 20 [cited 2019 Mar 13];10:205–8. Available from: https://dx.doi.org/10.2147/PGPM.S107152
- Leger P, Chirwa S, Nwogu JN, Turner M, Richardson DM, Baker P, et al. Race/ethnicity difference in the pharmacogenetics of bilirubin-related atazanavir discontinuation. Pharmacogenet Genomics [Internet]. 2018 Jan [cited 2019 Mar 13];28(1):1–6. Available from: http://insights.ovid.com/pubmed?pmid=29117017
- Johnson DH, Venuto C, Ritchie MD, Morse GD, Daar ES, McLaren PJ, et al. Genomewide association study of atazanavir pharmacokinetics and hyperbilirubinemia in AIDS Clinical Trials Group protocol A5202. Pharmacogenet Genomics [Internet]. 2014 Apr [cited 2018 Jul 20];24(4):195–203. Available from: http://insights.ovid.com/pubmed?pmid=24557078
- Rodríguez-Nóvoa S, Martín-Carbonero L, Barreiro P, González-Pardo G, Jiménez-Nácher I, González-Lahoz J, et al. Genetic factors influencing atazanavir plasma concentrations and the risk of severe hyperbilirubinemia. AIDS [Internet]. 2007 Jan 2 [cited 2018 Jul 20];21(1):41–6. Available from: http://insights.ovid.com/pubmed?pmid=17148966
- Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmöller J, Johne A, et al. Functional polymorphisms of the human multidrug- resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A [Internet]. 2000 Mar 28 [cited 2019 Mar 13];97(7):3473–8. Available from: http://www.pnas.org/cgi/pmidlookup?view=long&pmid=10716719
- Rotger M, Taffe P, Bleiber G, Gunthard HF, Furrer H, Vernazza P, et al. Gilbert syndrome and the development of antiretroviral therapy-associated hyperbilirubinemia. J Infect Dis [Internet]. 2005 Oct 15 [cited 2019 Mar 10];192(8):1381–6. Available from: https://academic.oup.com/jid/article-lookup/doi/10.1086/466531
- Boyd MA, Srasuebkul P, Ruxrungtham K, Mackenzie PI, Uchaipichat V, Stek MJr, et al. Relationship between hyperbilirubinaemia and UDP-glucuronosyltransferase 1A1 (UGT1A1) polymorphism in adult HIV-infected Thai patients treated with indinavir. Pharmacogenet Genomics [Internet]. 2006 May [cited 2019 Mar 13];16(5):321–9. Available from: http://insights.ovid.com/pubmed?pmid=16609363
- Fukuen S, Fukuda T, Maune H, Ikenaga Y, Yamamoto I, Inaba T, et al. Novel detection assay by PCR-RFLP and frequency of the CYP3A5 SNPs, CYP3A5*3 and *6, in a Japanese population. Pharmacogenetics [Internet]. 2002 Jun [cited 2019 Mar 13];12(4):331–4. Available from: http://insights.ovid.com/pubmed?pmid=12042671
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet [Internet]. 2001 Apr [cited 2019 Mar 13];27(4):383–91. Available from: http://dx.doi.org/10.1038/86882
- Saitoh A, Singh KK, Powell CA, Fenton T, Fletcher CV, Brundage R, et al. An MDR1-3435 variant is associated with higher plasma nelfinavir levels and more rapid virologic response in HIV-1 infected children. AIDS [Internet]. 2005 Mar 4 [cited 2018 Jul 20];19(4):371–80. Available from: http://insights.ovid.com/pubmed?pmid=15750390
- Hartkoorn RC, Kwan WS, Shallcross V, Chaikan A, Liptrott N, Egan D, et al. HIV protease inhibitors are substrates for OATP1A2, OATP1B1 and OATP1B3 and lopinavir plasma concentrations are influenced by SLCO1B1 polymorphisms. Pharmacogenet Genomics [Internet]. 2010 [cited 2019 Mar 13];20(2):112–20. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4859410/pdf/emss-30396.pdf
- Michaud V, Bar-Magen T, Turgeon J, Flockhart D, Desta Z, Wainberg MA. The dual role of pharmacogenetics in HIV treatment: mutations and polymorphisms regulating antiretroviral drug resistance and disposition. Pharmacol Rev [Internet]. 2012 Jul [cited 2019 Mar 12];64(3):803–33. Available from: http://pharmrev.aspetjournals.org/cgi/pmidlookup?view=long&pmid=22759796
- Kohlrausch FB, de Cassia Estrela R, Barroso PF, Suarez-Kurtz G. The impact of SLCO1B1 polymorphisms on the plasma concentration of lopinavir and ritonavir in HIV-infected men. Br J Clin Pharmacol [Internet]. 2010 Jan [cited 2019 Mar 12];69(1):95–8. Available from: https://doi.org/10.1111/j.1365-2125.2009.03551.x
- Tarr PE, Taffé P, Bleiber G, Furrer H, Rotger M, Martínez R, et al. Modeling the influence of APOC3, APOE, and TNF polymorphisms on the risk of antiretroviral therapy-associated lipid disorders. J Infect Dis [Internet]. 2005 May 1 [cited 2019 Mar 13];191(9):1419–26. Available from: https://academic.oup.com/jid/article-lookup/doi/10.1086/429295
- Arnedo Valero M, Taffé P, Sahli R, Furrer H, Hirschel B, Elzi L, et al. Contribution of 20 variants of 13 genes to dyslipidemia associated with antiretroviral therapy. Pharmacogenet Genomics [Internet]. 2007 Oct [cited 2018 Jul 20];17(9):755–64. Available from: https://www.researchgate.net/publication/6140058_Contribution_of_20_single_nucleotide_polymorphisms_of_13_genes_to_dyslipidemia_associated_with_antiretroviral_therapy
- Foulkes A. Protease inhibitors, Apolipoprotein C-III polymorphisms and atherogenic dyslipidemia in HIV. Presentation at the12th Conference on Retroviruses and Opportunistic Infections; Boston; 2005.
- Tsuchiya K, Hayashida T, Hamada A, Oki S, Oka S, Gatanaga H. High plasma concentrations of dolutegravir in patients with ABCG2 genetic variants. Pharmacogenet Genomics [Internet]. 2017 Nov [cited 2019 Mar 13];27(11):416–9. Available from: http://insights.ovid.com/pubmed?pmid=28858994
- Martínez Rodríguez A, Duque Vizcaíno M, Castro Peraza O, Alfonso Orta I, Jiménez López G, Acosta Reynaldo ED. Notificaciones espontáneas de sospechas de reacciones adversas a los medicamentos. Instituto “Pedro Kourí”. Rev Cubana Farm [Internet]. 2016 [cited 2019 Mar 13];50(1). Available from: http://www.revfarmacia.sld.cu/index.php/far/article/view/6. Spanish.
- Álvarez Barco E, Rodríguez Nóvoa S. The Pharmacogenetics of HIV Treatment: A Practical Clinical Approach. J Pharmacogenom Pharmacoproteomics [Internet]. 2013 [cited 2018 Jul 18];4(1). Available from: https://www.longdom.org/open-access/the-pharmacogenetics-of-hiv-treatment-a-practical-clinical-approach-2153-0645.1000116.pdf
- Haas D, Anderson J, Sanne I, Wilkinson G, Quinn J. Pharmacogenetics of nevirapine (NVP) and hepatotoxicity: ans AACTG collaborative study. Presentation at the 12th Conference on Retroviruses and Opportunistic Infections; Boston; 2005.
- FDA [Internet]. Maryland: U.S. Food & Drug Administration; c2019. Science & Research Drugs. Table of Pharmacogenomic Biomarkers in Drug Labeling; 2018 [updated 2018 Jun; cited 2019 Mar 9]. Available from: https://www.fda.gov/drugs/scienceresearch/ucm572698.htm
- Chen S, St Jean P, Borland J, Song I, Yeo AJ, Piscitelli S, et al. Evaluation of the effect of UGT1A1 polymorphisms on dolutegravir pharmacokinetics. Pharmacogenomics [Internet]. 2014 [cited 2019 Mar 13];15(1):9–16. Available from: https://www.futuremedicine.com/doi/10.2217/pgs.13.190?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dwww.ncbi.nlm.nih.gov
- Center for State Control of Medicines, Equipment and Medical Devices (CECMED). Guía para la realización de los estudios farmacogenómicos durante el desarrollo de los medicamentos. [Internet]. Havana: CECMED; 2018 [updated 2018 Apr 18; cited 2019 Mar 13]. 16 p. Available from: https://www.cecmed.cu/sites/default/files/adjuntos/Reglamentacion/guia_farmacogenomico.pdf. Spanish.
- Kiser J, Aquilante C, Anderson P, Wolfe P, Carten M, Fletcher C. Effect of multidrug resistance proteins 2 and 4 polymorphisms on tenofovir pharmacokinetics in HIV-infected patients. Presentation at the 7th International Workshop on Clinical Pharmacology of HIV Therapy; Lisbon, Portugal; 2006.
- Arab-Alameddine M, Di Iulio J, Buclin T, Rotger M, Lubomirov R, Cavassini M, et al. Pharmacogenetics-based Population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Swiss HIV CohortStudy. Clin Pharmacol Ther [Internet]. 2009 May [cited 2019 Mar 13];85(5):485–94. Available from: https://doi.org/10.1038/clpt.2008.271
- Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS [Internet]. 2009 Oct 23 [cited 2019 Mar 13];23(16):2101–6. Available from: http://insights.ovid.com/pubmed?pmid=19779319
- Fauvel J, Bonnet E, Ruidavets JB, Ferrières J, Toffoletti A, Massip P, et al. An interaction between apo C-III variants and protease inhibitors contributes to high triglyceride/low HDL levels in treated HIV patients. AIDS [Internet]. 2001 Dec 7 [cited 2019 Mar 5];15(18):2397–406. Available from: http://insights.ovid.com/pubmed?pmid=11740190
THE AUTHORS
Danays García-Blanco (Corresponding author: danaysgb@ipk.sld.cu), pharmacist, Pharmacological Research Department, Pedro Kourí Tropical Medicine Institute (IPK), Havana, Cuba.
Rosario Gravier-Hernández, biochemist with a master’s degree in pharmacology, Pharmacological Research Department, IPK, Havana, Cuba.
Carlos L. Rabeiro-Martínez, pharmacist with a master’s degree in pharmacology, Pharmacological Research Department, IPK, Havana, Cuba.
Lizette Gil del Valle, biochemist with a doctorate in pharmaceutical sciences. Associate professor, Head of Pharmacological Research Department, IPK, Havana, Cuba.
Jorge Pérez-Ávila, physician, specializing in pharmacology, with a master’s degree in pharmacology. Consulting professor, IPK, Havana, Cuba





