INTRODUCTION
The use of highly active antiretroviral treatment (HAART) has reduced progression to AIDS and remarkably increased survival in HIV-positive patients. These benefits are threatened by emergence of antiviral resistance—due to mutations in HIV genes coding for viral proteins targeted by antiretroviral (ARV) drugs—resulting in partial to total loss of susceptibility to inhibitors, impeding their therapeutic effects.[1]
Mother-to-child transmission (MTCT) of HIV-1 is the main cause of pediatric HIV-1 infection and may occur in utero, during delivery or with breastfeeding.[2] Worldwide, about 1600 children are infected daily, 90% of them in Sub-Saharan Africa, where HIV-1 MTCT is the main contributor to infant mortality.[3]
Earlier, only sporadic reports appeared in the literature describing MTCT by drug-resistant HIV-1 strains.[4] However, more recent studies have found that 9% to 30% of HIV-1–infected children were born to mothers who received prophylactic zidovudine (AZT), demonstrating resistance to the drug among these newborns.[5–10] Thus, while antiretroviral therapy (ART) has led to important advances in the last decade in preventing MTCT HIV-1 transmission, there remain some situations that are particularly risky: unknown maternal HIV status, primary HIV infection during pregnancy, and suboptimal maternal ART prophylaxis, among others.[10,11]
Since the efficacy of AZT was first demonstrated in 1994, a gradual decrease of vertical HIV transmission has been observed in Europe and the United States, reducing rates from 25% to below 2%.[10,12,13] Based on such evidence, all newborns of HIV-1 seropositive mothers in Cuba are treated with AZT in the first six weeks after birth. If HIV-1 infection is confirmed, they are started on ARV treatment.
Domestically-manufactured ARVs have been used in Cuba since 2001. At first, limited supplies meant only some patients could be treated; children and pregnant women were among those given priority.[14] ARV drugs most frequently used are: nucleoside reverse transcriptase inhibitors (NRTI)—lamivudine (3TC), stavudine (D4T) and zidovudine (AZT); non-nucleoside reverse transcriptase inhibitors (NNRTI)—nevirapine (NVP); and protease inhibitors (PI)—indinavir (IDV). The most frequently used regimen includes 3TC, AZT and NVP. The most widely used antiretroviral in combined treatments is 3TC.[14]
Until 2008, all HIV-positive pregnant women in Cuba were treated with AZT beginning at 14 weeks of gestation; and depending on immune status, additionally with 3TC and NVP. At the end of 2008, the therapeutic protocol was changed, introducing the triple therapy used today (3TC, AZT and NVP). When HIV infection is diagnosed late (during third trimester of pregnancy), AZT, 3TC and a lopinavir/ritonavir (Kaletra) booster are used.[15]
By the end of 2009, 12,217 persons had been diagnosed with HIV-1 infection in Cuba. Of 402 children born to HIV-1–positive mothers, 36 were infected by vertical transmission (9%) and five (1.2%) through blood. At the time of this study, 25 children were alive and 22 were receiving ART (National HIV Registry 2009 Annual Report, Ministry of Public Health, Havana).
Previous molecular epidemiology reports on HIV-1 describing most frequent subtypes and ARV resistance have been limited to adult populations.[9–15]
The objective of this study is to identify ARV-resistant HIV-1 strains in Cuban children and their mothers; and to do a phylogenetic characterization and comparison of HIV-1 pol gene sequences in the same population.
METHODS
A descriptive retrospective study was carried out of Cuban HIV-1–infected children and their mothers. The study universe was the 25 HIV-1−infected children alive at the end of 2009, and their mothers.
Inclusion criteria Eligible were mother–child pairs with children aged <18 years treated in the Pedro Kourí Tropical Medicine Institute (IPK, its Spanish acronym) pediatric clinic in Havana from 2004 through 2009. This resulted in a study sample of 22 pairs. Children’s age ranged from 11 months to 5 years.
Ethics The study objective was explained to all adults, who provided written informed consent for their enrollment. The study protocol was approved by the IPK ethics committee, and was designed in accordance with the Helsinki Declaration.
Samples for laboratory assay Blood samples were drawn in the IPK clinical laboratory. Second samples were collected from 11 children and a third sample from one boy, for a total of 56 samples received and processed. Five milliliters of whole blood were collected in tubes anticoagulated with EDTA. Blood was centrifuged at 1000 gravities (g) for 15 minutes to obtain 2 mL of plasma, which was stored at −70 oC until processing.
Extraction of virus RNA HIV RNA genome was obtained from 1 mL of plasma by centrifugation at 14,000 g for 1 hour at 4 oC. The supernatant was discarded, leaving a volume of 140 µL, in which the virus precipitate was resuspended. Virus RNA was extracted from those 140 µL using the QIAamp Viral RNA Mini commercial kit (QIAGEN, Germany) following the manufacturer’s method. Vials with extracted RNA were stored at −70 oC until use.
Reverse transcription and amplification with polymerase chain reaction (PCR) Reverse transcription (conversion of the HIV RNA molecule into a DNA copy) followed by PCR amplification of 2060 bp of the HIV pol gene was carried out using the commercial kit SuperScript One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen, USA) under the following conditions: 15 µL of 2X reaction buffer (0.4 mM of each of the deoxinucleotides, dNTP, and 2.4 mM MgSO4), 0.5 µL of each primer (av159 and av192) at 10 uM and 8 µL of 40 mM MgSO4, 1 µL of 50 U SuperScript III RT with Platinum Taq High Fidelity enzyme supplemented with 0.25 µL of 10 U RNAse protector (Roche, Germany) and the 40 µL volume was completed with water. Finally, 10 µL of extracted RNA were added to this mixture, for a final reaction volume of 50 µL.
Reverse transcription took place at 55 oC for 30 minutes, followed by an initial denaturation at 94 oC for 2 minutes and 40 cycles of: denaturation at 94 oC for 15 seconds, hybridization at 61 oC for 30 seconds, and elongation at 68 oC for 3 minutes. A last elongation step took place at 68 oC for 5 minutes.
A second polymerase chain reaction (nested PCR) was performed to obtain a 1745 bp internal fragment of the pol gene using the Expand High Fidelity PCR System commercial kit (Roche, Germany). Two tubes were prepared for each sample to carry out this reaction. The first tube contained a mixture of 0.4 µL of 10mM dNTPs, 1.25 µL of each primer (av190 and av191) at 2.5 mM concentration, and water to complete a volume of 23 µL. The second tube contained a mixture prepared with 5 µL 10X Buffer Expand, 6 µL of 150 mM MgCl2, 0.75 µL of 2.625 U Expand HF Enzyme (Roche, Germany), and water to complete a volume of 25 µL. Two milliliters of amplified cDNA (complementary DNA, synthesized by reverse transcriptase from a Messenger RNA template) were then added to the first tube, for a total volume of 25 µL. The second mixture was added to the first for a final reaction volume of 50 µL. This reaction was subjected to 94 °C for 2 minutes and then 30 cycles of: 94 °C for 15 seconds, 59 °C for 30 seconds, and 68 °C for 3 minutes. Finally there was an elongation at 72 °C for 10 minutes.
Purification and sequencing Ten microliters of the nested PCR product were used, plus 2 µL of run indicator (Blue/Orange 6 X loading Dye, Promega, USA) and applied to a 1% agarose gel stained with ethidium bromide (BDH, 0.1 µg/mL) to perform horizontal electrophoresis using a DNA standard of 1 kb molecular weight (Step Ladder, Promega, USA). Presence of amplified virus DNA was detected by observing the electrophoresis-separated samples through a UV transilluminator. A visible band should show of the size expected (1745 bp). The electrophoresis-positive PCR products were subjected to column purification using the QIAquickPCR Purification (QIAGEN, Germany) kit following manufacturer’s instructions.
To obtain both sense nucleotide sequences of the pol gene 1302 bp fragment—which codes for the HIV-1 99 protease amino acids and 335 reverse transcriptase amino acids—6 sequence reactions were performed with different primers for each purified DNA (KVL162, KVL163, KVL164, KVL165, AV5, KVL176). For the sequencing reactions, mixtures formed by 1 µL of each sequencing primer, 8 µL of sequence reaction mixture DTCS Quick Star Master Mix (supplied with the Dye Terminator Cycle Sequencing (DTCS) Quick Start Kit, from Beckman Coulter, USA), 5 µL of purified DNA (approximately 100 ng) and 6 µL of water to complete 20 µL of reaction mixture. The sequencing reaction consisted of two minutes denaturation at 96 ºC, followed by 50 cycles of: denaturing at 96 ºC for 20 seconds, 20 seconds of hybridization at 50 ºC and 4 minutes of extension at 60 ºC. Once the sequence reaction was concluded, it was purified, following the protocol described in the DTCS Quick Star Master Mix commercial kit (Beckman Coulter, USA). The sequencing reaction run was done in a Beckman Coulter model CEQ8800 (USA automatic sequencer), using the crude data analysis procedure for PCR products.
The primers to amplify and sequence the corresponding HIV-1 pol gene region were designed by Leathem and Vandamme at the Rega Medical Research Institute in Belgium, using OLIGO 5.0, 2007 (Personal Communication: Kristel Van Leathem and Anne Mieke Vandamme, Rega Institute for Medical Research, Louvain Catholic University, Belgium).
Sequence edition The six sequence reactions for the pol gene fragment obtained from each purified HIV-1 DNA, were assembled and edited with the Sequencher Version 4.9 (Gene Codes Corporation, USA) platform, using as a reference the nucleotide sequence of HIV-1 strain B.FR.83.HXB2_LAI_IIIB_BRU.K03455, to obtain a single HIV-1 consensus sequence for every patient’s virus.
Phylogenetic analysis To define HIV-1 subtypes or genetic variants in samples studied and similarity or divergence between pairs, phylogenetic analysis constructing trees by neighbor joining was performed based on Kimura’s method for distance between two parameters[16] using the MEGA program version 4.[17] The analysis of recombinant genetic forms was done by bootscanning [18], using the Simplot V, version 3.5.1.[19] Tree topology reproducibility was evaluated by bootstrapping using 1000 replicas. A phylogenetic group was defined as having a bootstrap value of ≥70%.[20] Evolutionary distances were estimated with the DNADist program using Kimura’s two parameter method.[16]
Mutation analysis and resistance inference The complete 99 amino-acid protease (PR) sequence and 335 amino-acid sequence of reverse transcriptase (RT) of each previously sequenced and edited virus was sent to the Stanford University database to determine mutations conferring resistance to transaminase and protease inhibitors, as well as resistance levels (high, intermediate, low or susceptible) to each specific drug.[21]
RESULTS
Of all samples processed, HIV amplification was achieved in only 15 of the children’s samples (11 children) and in 8 maternal samples (Table 1). This was probably because the viral load in negative cases was under 1000 copies/mL—the sensitivity limit for the PCR used to amplify the HIV-1 pol gene fragment in patients with ARV therapy failure.
Five children and one mother with viral amplification had received ARV; the mother had received AZT monotherapy. One child was sexually infected and the rest, vertically (transplacentally, perinatally or from breastfeeding) according to clinical records. Serial HIV specimens from three children were amplified and sequenced; three in Patient 4 (sequences NCB4, NCB4a and NCB4b), and two samples from the other two, Patient 1 (sequences NCB1 and NCB1a) and Patient 2 (sequences NCB2 and NCB2a) (Table 1).
The sample was too small to permit statistical inferences for proportions, subtypes, or mutation development.
Phylogeny Subtype analysis showed recombinant form CRF19_cpx present in four children; subtype B in three; CRF18_cpx, in two and subtype C in one. All viruses obtained in children of mother−child pairs were coincident for one subtype, results supported by high bootstrapping values observed in trees (≥75%) (Figure 1; Table 2).
Estimated evolutionary divergence between sequences in mother–child dyads MCB1–NCB1, MCB4–NCB4, MCB5–NCB5, and MCB9–NCB9 is almost zero, with values of 0.008, 0.002, 0.003, and 0.002, respectively. This is because the samples were obtained within two years of diagnosis; moreover, in all four pairs the dates of diagnosis were within five weeks of one another. The greatest divergence values were found between MCB7–NCB7 (0.010) and MCB11–NCB11 (0.082). Samples from these patients were collected several years after HIV infection diagnosis, which could affect the values obtained (Table 1).
Estimated evolutionary divergence between sequences of HIV-1 serial specimens of the same patient was zero (0.000) for NCB1 and NCB1a and for NCB2 and NCB2a; that is, serial sequences of these two children were almost identical. However, between the first NCB4 virus and the third NCB4b, this value was 0.012; while between the second (NCB4a) and third (NCB4b) the value was 0.008, demonstrating that this child’s virus changed over time. Of the serial specimens from Patient 4 (NCB4, NCB4a y NCB4b), the second one was obtained three months later, when the child was already in ARV therapy, and two mutations were detected in the virus. The third sample was collected 33 months after the first; by that time, a larger number of mutations had accumulated, presenting even greater divergence between the first and last samples.
HIV-1 mutations associated with ARV resistance Treated children Mutations conferring resistance to ARV were detected in viruses of all five children under treatment at time of sample collection (NCB1, NCB2, NCB4, NCB8 and NCB11). In patients with serial plasma samples (seven samples), mutations were detected in more than one sample (Table 2).
Four children were being treated with 3TC, NVP, D4T and one with 3TC, NVP, AZT, corresponding to viral mutations and resistances found (Table 2).
The most frequent mutations were M184V (conferring high resistance to 3TC and FTC medications), found in five samples from three patients and K103N (conferring resistance to NNRTIs), detected in six samples from four patients. Mutations K70KN, F116Y, A98AG, V108IV, L210F, H221HY, F227FL and G190AG, which affect sensitivity to NRTI and NNRTI drugs, were observed less frequently. Mutation Q151M, found in amplified virus of the third sample of Patient NCB4b is noteworthy, since it is accompanied by sequential accumulation of other mutations increasing loss of sensitivity to all NNRTI (Table 2).
In repeat samples from three patients (NCB1, NCB2, NCB4), viruses were detected that maintained resistance mutations from the first sample. This is because treatments were not changed, either because patients did not fulfill clinical, immunologic or virologic criteria for therapeutic failure; or because the country lacked genotype assays for assessing how best to manage these patients.
Untreated children Mutations conferring ARV resistance were detected in two of six viruses sequenced from children who had not been treated at the time of sampling (NCB3, NCB10). Mutations L90LM and V106IV were found in Patient NCB10’s virus. The former is associated with reduced PI sensitivity. The exact time this child was infected is unknown, but his mother was not infected at time of delivery, since she was tested twice during her pregnancy, both were diagnosed more than five years later, and he had been breastfed for three years. We therefore infer that this patient acquired HIV from breastfeeding and that his mother transmitted the resistant strain amplified in his sample, although it was not found in hers. The virus population detected in the child could have gone undetected in the mother if it was not the majority virus at the time of sample collection; minority virus populations (less than 20%) may not be detected by the usual techniques used for resistance studies.[22]
Patient NCB3’s virus had multiple mutations (M184V, T215Y, A98G, K103S and G190A) conferring resistance to NRTIs and NNRTIs (Table 2). This child was born to an HIV-infected mother under ARV therapy, so measures had been taken to avoid vertical transmission (birth was by caesarean section and the baby was not breastfed). Even so, the child was infected, probably because of the mother’s poor compliance with therapy (data from clinical records). There was no virus amplification in the mother’s sample, so we could not demonstrate presence of these mutations, but we infer that she transmitted this multi-resistant strain to her child, since she was treated in 2004 with 3TC, NVP and D4T, and in 2005 with 3TC, NVP y AZT.
Table 1: Information on patients whose samples achieved viral amplification
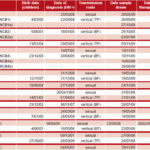
BF: Breastfeeding; TP: transplacental; NCB: Cuban child; MCB: Cuban mother; NT: not treated at time of sample Source: Clinical records, IPK
Child NCB7 did not show the V118I mutation detected in the mother (MCB7), possibly because four years had passed between the child’s birth and the time of sample collection.
Untreated mothers In the analysis of viruses obtained from untreated mothers, mutations were detected in two of eight patients (MCB7 and MCB12); one had the V118I mutation and the other, K219Q/E, both of which affect sensitivity to NRTIs in the presence of other mutations, but not on their own.
DISCUSSION
HIV evolutionary studies and detection of virus mutations conferring ARV resistance in the pediatric population require serial specimens from children from birth, and from mothers before starting treatment, at time of delivery and thereafter. Thus identifying resistant strains transmitted provides a timely and effective aid in clinical management.[23] Among the limitations of this study are the absence of viral amplification and lack of serial specimens from the women in pregnancy and from some of the children. Obtaining serial specimens from pregnant women is difficult, since most HIV-positive women decide to become pregnant when they are clinically, immunologically and virologically compensated and so have undetectable viral loads.
In the present study, most mothers in whose samples virus amplification was achieved were diagnosed at the end of pregnancy or after delivery—in some cases, more than a year later—infecting their children during breastfeeding. So in some cases we are missing the children’s samples at birth because they were not diagnosed until later; in some for whom we did have a sample taken at birth, the viral load was undetectable. Note that the PCR technique used in ARV resistance studies is not the same that is used for diagnosis and has different sensitivity limits.[24−27] The technique used in this study can detect viral loads between 500 and 1000 copies/mL.
All analyzed sequences of the children’s pol gene fragments coincided with those of their respective mothers and were consistent with subtypes previously seen in Cuba.[28–30] Evolutionary divergences between viruses obtained from mothers and their children were very small (in some cases zero), consistent with the short time elapsed between collection of samples from mother and child at the time of diagnosis. This was also true for children’s serial specimens. The greatest divergences occurred when mothers had been infected more than five years previously.[31,32]
Table 2: Treatment history, mutations and medications associated with resistance and virus subtype for patients whose samples achieved viral amplification

Clinical records; Stanford University Database
Most pediatric patients studied were infected by vertical transmission as a result of delayed diagnosis of HIV infection in the mother. Cuba has reported reduced rates of vertical transmission with correct ARV compliance in the infected mother.[33,34] However, possibly due to inadequate ARV compliance, we have detected ARV resistance mutations which may then be transmitted from mother to child.[35−37] Both timely diagnosis and more powerful therapy are increasingly important,[38] which is why Cuba changed its treatment protocol for HIV-1–infected pregnant women in 2008.[15]
Figure 1: Phylogenetic trees (by neighbor-joining) of 23 hiv-1 pol gene sequences from mothers and children

Analyses are presented in a single tree, corresponding to sequences clustering with HIV-1 subtypes and recombinant forms Subtype B, CRF19_cpx, Subtype C and CRF18_cpx. Trees were constructed using Kimura’s two-parameter method; shown for bootstrapping values ≥70%. Viruses corresponding to subtypes and reference CRF are underlined and denoted by subtype or corresponding CRF followed by isolate name. Viruses designated as NCB (children) or MCB (mothers) belong to this study and are preceded by a triangle ▲ (NCB) or circle ● (MCB).
It is critical that resistance assays be done before starting ARV. Therapeutic options for children are already limited because not all drugs are available in pediatric formulations.[39–41] Vertically-transmitted resistances further limit therapeutic options.[39,42] Delaugerre and colleagues describe two mechanisms involved in MTCT of resistant strains. The first involves a majority population in the mother transmitted to the child during delivery; the second, a minority-resistant population that becomes majority in the child.[10] As is seen in this study, in some cases MTCT of resistant viruses can be inferred, if not proven, since some mutations are observed in children that for various reasons are not found in their mothers.
Since commencing domestic ARV manufacture, Cuba’s health system has given priority to mothers and children.[14] However, mutations have been found in all children treated, sometimes after as little as 4 months’ therapy. This underscores the need for resistance assays in pregnant and puerperal women and in the infected child to search for transmitted resistant strains. Early appearance of mutations may result from the use of low genetic barrier medications, for which just one mutation can cause loss of sensitivity, or may occur when an undetected minority-viral population rapidly emerges in the presence of an antiviral to which it is resistant.[43]
Rapid emergence of resistance to 3TC and NVP with the appearance of M184V and K103N mutations, respectively, has been reported and worldwide.[40,44–47] In our pediatric patients under treatment, we also found the NRTI-resistant HIV-1 mutations most commonly found by Adjé-Touré et al.[41] Gupta et al. also found these to be the most frequent mutations, due to first-line use of NRTI.[40]
Our results for untreated patients are consistent with those of Simonetti et al., who detected virus mutations conferring resistance more frequently in treated than in untreated populations.[48] The mutations detected in untreated mothers (V118I and K219Q/E) only affect sensitivity to NRTIs in the presence of other mutations and may be a natural result of the high variability characteristic of HIV.[49]
Pediatric treatment of HIV infection has the same goals regardless of context: to limit vertical transmission;[38,39] to minimize emergence of resistant viruses in mother and child if vertical transmission does occur; and in such cases, to characterize the viral ARV resistance profile in the child beginning treatment.[23] Without resistance tests, effective first-line therapy is essential.
CONCLUSIONS
All sequences of the HIV-1 pol gene obtained from children in this study were the same as those of their respective mothers and are among the subtypes previously seen in Cuba. We detected mutations conferring resistance to NRTI in all amplified viruses from treated children, consistent with the drugs used in that population. In samples taken from mothers and children at the time of diagnosis, we detected possible resistant-strain transmission. The results of this study highlight the importance of resistance assays in pediatric HIV patients at the time of diagnosis prior to starting therapy, and, if treatment failure is suspected, during therapy. Strict adherence to ARV protocols is critical in both pregnant women and children, since early infection with resistant strains can have dire long-term consequences for both mother and child, undermining treatment effectiveness.
This study is the first in Cuba to determine levels of ARV-resistant viruses in HIV-1–infected mothers and children, describe related mutations, and correlate them with therapy used. The results of this study make an important contribution to patient management and selection of appropriate therapy and follow-up, helping improve their prognosis and quality of life.






